Documentation/3.6
From Slicer Wiki
Home < Documentation < 3.6
This is work in progress
Contents
Main GUI
- Main Application GUI
- "Hot-keys" and Keyboard Shortcuts
- Loading Scenes and Individual Datasets through the Data Module
- Data Loading Details
- Saving Scenes and Data
- Creating and Restoring Scene Snapshots
- Extensions Management Wizard
Modules
- Please copy the template linked below, paste it into your page and customize it with your module's information.
Slicer3:Module_Documentation-3.5_Template Not yet 3.6
- See Requirements for Modules for info to be put into the Help and Acknowledgment Tabs
- To put your lab's logo into a module, see here
Please adhere to the naming scheme for the module documentation:
- [ [Modules:MyModuleNameNoSpaces-Documentation-3.6|My Module Name With Spaces] ] (First Last Name)
Requirements for Modules
|
Examples for the Help and
Acknowledgment Panels |
List of Modules added since the 3.4 release
- Atlas Creator (Sylvain Jaume) Not yet 3.6
- MRI Bias Field Correction (Sylvain Jaume) Not yet 3.6
- 4D Image (Viewer) (Junichi Tokuda) Not yet 3.6
- 4D Analysis (Time-intensity curve plotting and analysis) (Junichi Tokuda) Not yet 3.6
- Fast Marching segmentation (Andriy Fedorov) Not yet 3.6
- Gyri Contour Segmentation (Peter Karasev) Not yet 3.6
- Subvolume extraction with ROI widget (Andriy Fedorov) Not yet 3.6
- Registration Metrics (HD and DSC) (Haytham Elhawary) Not yet 3.6
- Measurements (rulers and angles) (Nicole Aucoin) Not yet 3.6
- Affine registration (Casey Goodlett) Not yet 3.6
- Collect Patient Fiducials (Andrew Wiles) Not yet 3.6
List of pre-existing Modules
Core
- Camera Module (Sebastian Barre) Not yet 3.6
- Welcome Module (Wendy Plesniak, Steve Pieper, Sonia Pujol, Ron Kikinis) Not yet 3.6
- Volumes Module (Alex Yarmarkovich, Steve Pieper) Not yet 3.6
- Diffusion Editor (Kerstin Kessel) Not yet 3.6
- Models Module (Alex Yarmarkovich) Not yet 3.6
- Fiducials Module (Nicole Aucoin) Not yet 3.6
- Data Module (Alex Yarmarkovich) Not yet 3.6
- Slices Module (Jim Miller) Not yet 3.6
- Color Module (Nicole Aucoin) Not yet 3.6
- Interactive Editor (Steve Pieper) Not yet 3.6
- ROI Module (Alex Yarmarkovich) Not yet 3.6
- Volume Rendering Module (Yanling Liu, Alex Yarmarkovich) Not yet 3.6
Specialized Modules
Please adhere to the naming scheme for the module documentation:
- [[Modules:MyModuleNameNoSpaces-Documentation-3.6|My Module Name With Spaces] ] (First Last Name)
Wizards
- ChangeTracker (Andriy Fedorov) Not yet 3.6
- IA FE Meshing Module (Vince Magnotta) Not yet 3.6
Informatics Modules
- Fetch Medical Informatics Module (Wendy Plesniak) Not yet 3.6
- QDEC Module (Nicole Aucoin) Not yet 3.6
- Query Atlas Module (Wendy Plesniak) Not yet 3.6
Registration
- Overview of all Registration Modules
- Register Images (Upgraded Version) (Casey Goodlett, Stephen Aylward) Not yet 3.6
- Register Images Multires (Casey Goodlett, Stephen Aylward) Not yet 3.6
- Fiducial-Based Registration (Casey Goodlett) Not yet 3.6
- Transforms Module (Alex Yarmarkovich) Not yet 3.6
- Linear Registration (Daniel Blezek) Not yet 3.6
- Affine Registration (Daniel Blezek) Not yet 3.6
- Deformable B-Spline Registration (Bill Lorensen) Not yet 3.6
- Demons Registration (Tom Vercauteren) Not yet 3.6
- ACPC Transform (Nicole Aucoin) Not yet 3.6
Segmentation
- EM Segment (Sylvain Jaume, Nicolas Rannou) Not yet 3.6
- EM Segment Command-Line (Brad Davis, Will Schroeder) Not yet 3.6
- EM Segment Simple (Brad Davis, Will Schroeder) Not yet 3.6
- EM Segment Template Builder (Brad Davis, Will Schroeder) Not yet 3.6
- Simple Region Growing (Jim Miller) Not yet 3.6
- Otsu Threshold (Bill Lorensen) Not yet 3.6
Statistics
- Label Statistics (Steve Pieper) Not yet 3.6
Diffusion
DWI
- Estimation
- Diffusion Tensor Estimation (Raul San Jose Estepar) Not yet 3.6
- Python Extract Baseline DWI Volume (Julien de Siebenthal) Not yet 3.6
- Filter
- Joint Rician LMMSE Image Filter (Antonio Tristán Vega, Santiago Aja-Fernandez) Not yet 3.6
- Rician LMMSE Image Filter (Antonio Tristán Vega, Santiago Aja-Fernandez, Marc Niethammer) Not yet 3.6
- Unbiased Non Local Means filter for DWI (Antonio Tristán Vega, Santiago Aja-Fernandez) Not yet 3.6
- Python Shift DWI Values (Julien de Siebenthal) Not yet 3.6
- Python Recenter Scalar to DWI Volume (Julien de Siebenthal) Not yet 3.6
DTI
- Resample DTI Volume (Francois Budin) Not yet 3.6
- Diffusion Tensor Scalar Measurements (Raul San Jose Estepar) Not yet 3.6
- Analysis
Tractography
- Label Seeding (Raul San Jose Estepar) Not yet 3.6
- Fiducial Seeding (Alex Yarmakovich, Steve Pieper) Not yet 3.6
- FiberBundles (Alex Yarmakovich) Not yet 3.6
- Python Stochastic Tractography (Julien de Siebenthal) Not yet 3.6
- ROI Select (Lauren O'Donnell) Not yet 3.6
IGT
- OpenIGTLinkIF Module (Junichi Tokuda) Not yet 3.6
- NeuroNav Module (Haiying Liu) Not yet 3.6
- ProstateNav Module (Junichi Tokuda) Not yet 3.6
Filtering
- Checkerboard Filter (Bill Lorensen) Not yet 3.6
- Histogram Matching (Bill Lorensen) Not yet 3.6
- Image Label Combine (Alex Yarmarkovich) Not yet 3.6
- Resample Volume (Bill Lorensen) Not yet 3.6
- Resample Volume2 (Francois Budin) Not yet 3.6
- Threshold Image (Nicole Aucoin) Not yet 3.6
- Arithmetic
- Add Images (Bill Lorensen) Not yet 3.6
- Subtract Images (Bill Lorensen) Not yet 3.6
- Cast Image (Nicole Aucoin) Not yet 3.6
- Mask Image (Steve Pieper) Not yet 3.6
- Denoising
- Gradient Anisotropic Filter (Bill Lorensen checked this in) Not yet 3.6
- Curvature Anisotropic Diffusion (Bill Lorensen) Not yet 3.6
- Gaussian Blur (Julien Jomier, Stephen Aylward) Not yet 3.6
- Median Filter (Bill Lorensen) Not yet 3.6
- Morphology
- Voting Binary Hole Filling (Bill Lorensen) Not yet 3.6
- Grayscale Fill Hole (Bill Lorensen) Not yet 3.6
- Grayscale Grind Peak (Bill Lorensen) Not yet 3.6
Surface Models
- Modelmaker (Nicole Aucoin) Not yet 3.6
- Grayscale Model Maker (Bill Lorensen) Not yet 3.6
- Freesurfer Surface Section Extraction (Katharina Quintus)
- Python Surface Connectivity (Luca Antiga, Daniel Blezek) Not yet 3.6
- Python Surface ICP Registration (Luca Antiga, Daniel Blezek) Not yet 3.6
- Python Surface Toolbox (Luca Antiga, Daniel Blezek) Not yet 3.6
- Clip Model (Alex Yarmarkovich) Not yet 3.6
- Model into Label Volume (Nicole Aucoin) Not yet 3.6
Batch processing
- EM Segmenter batch (Julien Jomier, Brad Davis) Not yet 3.6
- Gaussian Blur batch (Julien Jomier, Stephen Aylward) Not yet 3.6
- Register Images batch (Julien Finet, Stephen Aylward) Not yet 3.6
- Resample Volume batch (Julien Finet) Not yet 3.6
Converters
- Create a Dicom Series (Bill Lorensen) Not yet 3.6
- Dicom to NRRD (Xiaodong Tao) Not yet 3.6
- Orient Images (Bill Lorensen) Not yet 3.6
- Python Explode Volume Transform (Luca Antiga, Daniel Blezek) Not yet 3.6
- Extract Subvolume (Steve Pieper) Not yet 3.6
Slicer Extensions
Extensions for Downloading
Introduction
- Slicer Extensions are a mechanism for third parties to provide modules which extend the functionality of 3d Slicer.
- Some of the extensions do not use the Slicer license. Please review carefully.
- For a subset of extensions, you can use the extension wizard in Slicer to find their webpages and to install/uninstall individual extensions. In case of problems with those modules, please talk directly to the developers of the extensions.
- The version that is available through the extension manager is chosen by the developer of that extension
We are using NITRC as the primary repository for contributed extensions. As a general rule, we do not test the extensions ourselves. Use them at your own risk. Click here to see a listing of Slicer 3 extensions on NITRC.
To add extension modules to an installed binary of slicer:
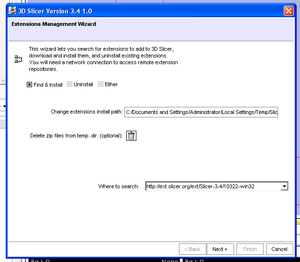
- Use the View->Extension Manager menu option
- The dialog will be initialized with the URL to the extensions that have been compiled to match your binary of slicer.
- Note installing extensions from a different repository URL is likely to be unstable due to platform and software version differences.
- You can select a local install directory for your downloaded extensions (be sure to choose a directory with enough free space).
- Select the extensions you wish to install and click to download them. Installed extensions will be available when you restart slicer.
- To turn modules on or off, you can use the Module Settings page of the View->Application Settings dialog.
- Extensions are compiled as part of the nightly build. In order to have your extension compiled nightly and made available to end users, please contact the Slicer team. For explanations for developers see here
Installation
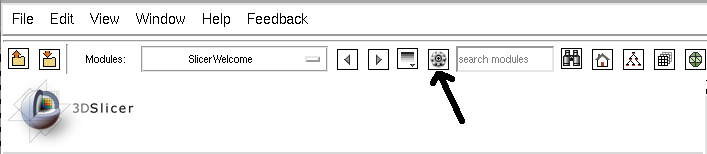
- Click on the icon to start the extensions wizard
Listing of available plug-ins
- VMTK (Daniel Haehn)
- You need to install the VmtkSlicerModule to run any of the other three. Not yet 3.6
- VMTKEasyLevelSetSegmentation - providing level-set segmentation of vessels, aneurysms and tubular structures using an easy interface Not yet 3.6
- VMTKLevelSetSegmentation - providing level-set segmentation of vessels, aneurysms and tubular structures using different algorithms Not yet 3.6
- VMTKVesselEnhancement - providing vessel enhancement filters to highlight vasculature or tubular structures Not yet 3.6
- EM DTI Clustering (Mahnaz Maddah) Not yet 3.6
- Label Diameter Estimation (Andriy Fedorov) Not yet 3.6
- ABC (Marcel Prastawa) Not yet 3.6
- Modules:HAMMER (GuorongWu, XiaodongTao, JimMiller, DinggangShen) Not yet 3.6
Extensions Contact List
This is a first pass list of extension authors; more to come!
| ABC | Marcel Prastawa | prastawa@sci.utah.edu |
| EMFiberClusteringModule | Mahnaz Maddah | maddah @nospam@ ge.com |
| BRAINSROIAuto | Greg Harris | gregory-harris @nospam@ uiowa.edu |
| Bubble Maker | Carlos Mendoza | carlos.sanchez.medoza @nospam@ gmail.com |
| Hammer Registration | Dinggang Shen | dgshen @nospam@ med.unc.edu |
| Lupus Lesion | Mark Scully | mscully @nospam@ mrn.org |
| VmtkSlicerModule | Daniel Haehn | haehn @nospam@ bwh.harvard.edu |
| Vmtkin3DSlicer | Daniel Haehn | haehn @nospam@ bwh.harvard.edu |
| BRAINSFit | Eun Young Kim | eunyoung-kim @nospam@ uiowa.edu |
| ARCTIC | Cedric Mathieu | ced.mathieu @nospam@ gmail.com |