Difference between revisions of "Announcements:Slicer3.6"
Sam.horvath (talk | contribs) Tag: 2017 source edit |
|||
| (74 intermediate revisions by 12 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | <noinclude>{{documentation/versioncheck}}</noinclude> | |
| − | |||
[[Documentation-3.6|Back to Documentation 3.6]] | [[Documentation-3.6|Back to Documentation 3.6]] | ||
| + | [[image:Slicer3-6Announcement-v2.png|3.6 Icon large]] | ||
=Introduction= | =Introduction= | ||
| Line 12: | Line 12: | ||
The main [http://www.slicer.org slicer.org] pages provide a guided tour to the application, training materials, and the development community. New users should start there because we try to keep the pages organized and up to date. | The main [http://www.slicer.org slicer.org] pages provide a guided tour to the application, training materials, and the development community. New users should start there because we try to keep the pages organized and up to date. | ||
| − | ==Highlights | + | |
| − | <gallery caption="Slicer v3.6 - New and Improved | + | =Our Philosophy= |
| − | Image: | + | Slicer is a community platform created for the purpose of subject specific image analysis and visualization. |
| − | Image:VolumeRenderingICPE.png| [[Modules:VolumeRendering-Documentation-3.6|Volume Rendering]] | + | |
| + | * Multi-modality imaging including, MRI, CT, US, nuclear medicine, and microscopy | ||
| + | * Multi organ from head to toe | ||
| + | * Bidirectional interface for devices | ||
| + | * Expandable and interfaced to multiple toolkits | ||
| + | |||
| + | There is no restriction on use, but permissions are responsibility of users. For details on the license see [http://www.slicer.org/cgi-bin/License/SlicerLicenseForm.pl here] | ||
| + | |||
| + | =Slicer Highlights= | ||
| + | <gallery caption="Slicer v3.6 - New and Improved Modules" widths="250px" heights="150px" perrow="3"> | ||
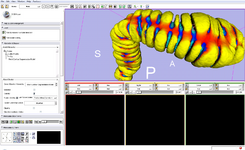
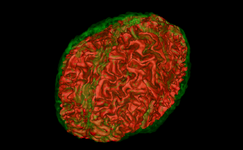
| + | Image:Slicer3.6-Editor-2010-05-06.png|The [[Modules:Editor-Documentation-3.6|Interactive Editor]] can be used to create and edit label maps for quantitative analysis and surface model generation (Steve Pieper) | ||
| + | Image:VolumeRenderingICPE.png| [[Modules:VolumeRendering-Documentation-3.6|Volume Rendering]] (Yanling Liu, Julien Finet, Lisa Avila) | ||
Image:Slicer-ColorGUI-3.6.jpg| [[Modules:Colors-Documentation-3.6|Colors]] (Nicole Aucoin) | Image:Slicer-ColorGUI-3.6.jpg| [[Modules:Colors-Documentation-3.6|Colors]] (Nicole Aucoin) | ||
| − | Image: | + | Image:Slicer3.6MeasurementsRulersModel.jpg| [[Modules:Measurements-Documentation-3.6 |Measurements (rulers and angles)]] (Nicole Aucoin) |
| + | Image:SlicerFiducialsGlyphs-3.4.jpg| [[Modules:Fiducials-Documentation-3.6|Fiducials]] (Nicole Aucoin) | ||
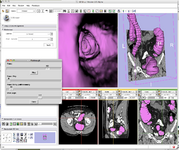
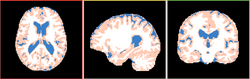
| + | Image:EMSegment-Simple.png|[[Modules:EMSegmenter-3.6|EM Segmenter]] (Kilian Pohl) | ||
Image:Fastmarching-result-adjusted.jpg| [[Modules:FastMarchingSegmentation-Documentation-3.6|Fast Marching segmentation]] (Andriy Fedorov) | Image:Fastmarching-result-adjusted.jpg| [[Modules:FastMarchingSegmentation-Documentation-3.6|Fast Marching segmentation]] (Andriy Fedorov) | ||
| − | Image: | + | Image:RSSkidneyL.png| [[Modules:RobustStatisticsSeg-Documentation-3.6|Robust Statistical Segmentation]] (Yi Gao) |
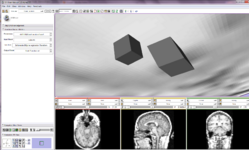
| − | Image: | + | Image:Fiducial registration before.jpg|NEW: [[Modules:RegisterImagesMultiRes-Documentation-3.6|Multiresolution Affine registration]] (Casey Goodlett) |
| − | Image: | + | Image:B-spline-reg-detail.png|Improved: [[Modules:RegisterImages-Documentation-3.6|Expert Automated Registration]] (Casey Goodlett) |
| − | Image: | + | Image:Fiducial_registration_before.jpg|NEW: [[Modules:TransformFromFiducials-Documentation-3.6|Fiducial Registration]] (Casey Goodlett) |
| + | Image:Registration_BRAINSfit.png|NEW: [[Modules:BRAINSFit |BRAINSFit Registration]] (Hans Johnson) | ||
| + | Image:Registration_BRAINSdemonwarp.png|NEW: [[Modules:BRAINSDemonWarp|BRAINSDemonWarp]] (Hans Johnson) | ||
| + | Image:BRAINSResample.png|NEW: [[Modules:BRAINSResample|BRAINSResample]] (Hans J. Johnson) | ||
| + | Image:BRAINSROIAuto.png|NEW: [[Modules:BRAINSROIAuto|BRAINSROIAuto]] (Hans J. Johnson) | ||
| + | Image:Plastimatch_icon.png|NEW: [[Modules:Plastimatch |Plastimatch non-rigid registration]] (Greg Sharp) | ||
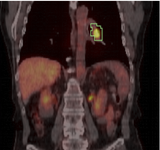
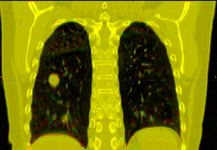
| + | Image:PETCTFusion.png| [[Modules:PETCTFusion-Documentation-3.6 |PETCTFusion]] (Wendy Plesniak) | ||
Image:| [[Modules:CollectFiducials-Documentation-3.6 |Collect Patient Fiducials]] (Andrew Wiles) | Image:| [[Modules:CollectFiducials-Documentation-3.6 |Collect Patient Fiducials]] (Andrew Wiles) | ||
Image:| [[Modules:IGTToolSelector-Documentation-3.6 |IGT Tool Selector]] (Andrew Wiles) | Image:| [[Modules:IGTToolSelector-Documentation-3.6 |IGT Tool Selector]] (Andrew Wiles) | ||
| − | Image: | + | Image:FourDImage Screenshot1.png| [[Modules:FourDImage-Documentation-3.6|4D Image Viewer]] (Junichi Tokuda) |
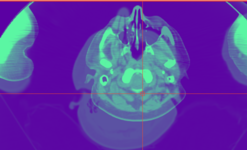
| − | Image | + | Image:MRI_Bias_Field_Correction_Slicer3_close_up.png|[[Modules:MRIBiasFieldCorrection-Documentation-3.6|MRIBiasFieldCorrection]] correction of MRI intensity inhomogeneity i.e. bias field (Sylvain Jaume) |
| − | Image: | ||
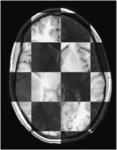
Image:N4 prostate bias ax.png| [[Modules:N4ITKBiasFieldCorrection-Documentation-3.6|N4 Bias Field Correction]] (Andriy Fedorov) | Image:N4 prostate bias ax.png| [[Modules:N4ITKBiasFieldCorrection-Documentation-3.6|N4 Bias Field Correction]] (Andriy Fedorov) | ||
| − | Image: | + | Image:Surface H c.png| [[Modules:MeshContourSegmentation-Documentation-3.6|Mesh Contour Segmentation]] (Peter Karasev) |
Image:ModelTransform.PNG| [[Modules:ModelTransform-Documentation-3.6|Model Transform]] (Alex Yarmarkovich) | Image:ModelTransform.PNG| [[Modules:ModelTransform-Documentation-3.6|Model Transform]] (Alex Yarmarkovich) | ||
Image:BrainWebImageTransformed.jpg| [[Modules:ResampleScalarVectorDWIVolume-Documentation-3.6|Resample Scalar/Vector/DWI Volume]] (Francois Budin) | Image:BrainWebImageTransformed.jpg| [[Modules:ResampleScalarVectorDWIVolume-Documentation-3.6|Resample Scalar/Vector/DWI Volume]] (Francois Budin) | ||
| + | Image:ExtractSubvolume3.png| [[Modules:CropVolume-Documentation-3.6|Crop Volume]] (Andriy Fedorov) | ||
| + | Image:Endoscopy-slicer3.6.png| [[Modules:Endoscopy-Documentation-3.6|Virtual Endoscopy]] (Steve Pieper) | ||
| + | Image:ChangeTracker_Analysis-3.4.png| [[Modules:ChangeTracker-Documentation-3.6|ChangeTracker]] (Andriy Fedorov) | ||
| + | Image:Slicer3_OpenIGTLinkIF__VolumeReslice2.png| [[Modules:OpenIGTLinkIF-Documentation-3.6|OpenIGTLinkIF]] (Junichi Tokuda) | ||
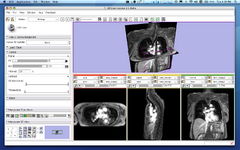
| + | Image:ProstateNav-3.6_Transrectal.png| [[Modules:ProstateNav-Documentation-3.6|ProstateNav]] (Andras Lasso, Junichi Tokuda) | ||
| + | Image:slicer3.6_neuronav_OR.jpeg| [[Modules:NeuroNav-Documentation-3.6|NeuroNav]] (Haiying Liu) | ||
| + | Image:IGTToolSelectorPointer.png |[[Modules:IGTToolSelector-Documentation-3.6|IGT tool selector]] (A. Wiles) | ||
| + | </gallery> | ||
| + | |||
| + | =Slicer Extensions= | ||
| + | <gallery caption="Slicer v3.6 - Extensions" widths="250px" heights="150px" perrow="3"> | ||
| + | Image:Plastimatch_dicomrt_dose.png|The [[Modules:PlastimatchDICOMRT| Plastimatch DICOM RT reader]] allows import and conversion of data in that format. (Greg Sharp) | ||
| + | Image:ABC out3d.png|The [[Modules:ABC-Documentation-3.5|ABC Segmenter]] is based on ITK EM technology. | ||
| + | Image:FCMResultLabels-3-6.png|[[Modules:FuzzySegmentationModule|Fuzzy segmentation]] | ||
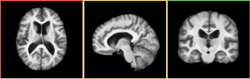
| + | Image:HammerAverage-3-6.png|[[Modules:HammerRegistration|Hammer Registration]] can be used to build statistical brain atlases | ||
| + | Image:Plastimatch image 2.png|[[Modules:Plastimatch|Plastimatch non-rigid registration]] (Greg Sharp) | ||
| + | Image:Corticalthickness.png|[[Modules:ARCTIC-Documentation-3.6|Arctic wizard]] (Automatic Regional Cortical ThICkness) | ||
| + | Image:LesionsWithVentricles.png|[[Modules:LesionSegmentationApplications-Documentation-3.6| Lupus white matter lesions segmentation]] (Jeremy Bockholt, Mark Scully) | ||
| + | Image:Shot2.png|[[Modules:EMDTIClustering-Documentation-3.6|EM DTI clustering]] (Mahnaz Maddah) | ||
| + | Image:RicianTensorCorrectionImage.png|[[Modules:RicianNoiseFilter|Rician Noise Filter]] for noise removal in DWI data | ||
| + | Image:FourDAnalysisModuleScreenShot-3.5.png|The [[Modules:FourDAnalysis-Documentation-3.6|FourD Analysis module]] was designed for time series analysis (Junichi Takuda) | ||
| + | Image:Avf 3d voronoi big.png|[[Modules:VMTKCenterlines|Centerline extraction using Voronoi diagrams]] (Luca Antiga, Daniel Haehn) | ||
| + | Image:Labeldiameterestimation1.png|[[Modules:LabelDiameterEstimation-Documentation-3.6|Label diameter estimation]] (Andriy Fedorov) | ||
| + | </gallery> | ||
| + | |||
| + | Checklist for the [[Checklist-3.6-Release|3.6 release]] | ||
| + | |||
| + | = Other Improvements, Additions & Documentation = | ||
| + | <gallery caption="" widths="250px" heights="150px" perrow="3"> | ||
| + | image:3.6 Default LUT.png| [[Slicer3:2010_GenericAnatomyColors|New default LUT]] (Michael Halle) | ||
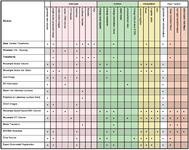
| + | Image:RegPortal_Icon.png|[[Slicer3:Registration|Registration Portal Page: overview of avail. tools & modules]] (Dominik Meier) | ||
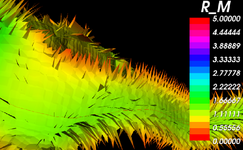
| + | Image:Resampling_DecisionMatrix.png|[[Registration:Resampling|Resampling Tool Feature Matrix]] (Dominik Meier) | ||
| + | Image:RegLib_Banner.png|[http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:UseCaseInventory Slicer Registration Case Library] (Dominik Meier) | ||
| + | Image:RegistrationWelcome_icon.png|[[Slicer3:Registration|Registration Welcome Module]] gives an overview of avail. registration modules | ||
| + | Image:SegmentationWelcome_icon.png|[[Modules:SegmentationOverview3.6|Segmentation Welcome Module]] gives an overview of avail. segmentation modules | ||
| + | Image:DiffusionWelcome_icon.png|[[Modules:DiffusionMRIWelcome-Documentation-3.6|Diffusion Welcome Module]] gives an overview of avail. DWI processing modules | ||
| + | Image:ITK_logo.png|[http://www.kitware.com/news/home/browse/272 ITK upgrades] provided performance improvements in all registration modules and many other modules. | ||
</gallery> | </gallery> | ||
Latest revision as of 21:14, 19 January 2022
Home < Announcements:Slicer3.6
|
For the latest Slicer documentation, visit the read-the-docs. |
Back to Documentation 3.6
Contents
Introduction
The community of Slicer developers is proud to announce the release of Slicer 3.6.
- Click here to download different versions of Slicer3 and find pointers to the source code, mailing lists and bug tracker. *Please note that Slicer continues to be a research package and is not intended for clinical use. Testing of functionality is an ongoing activity with high priority, however, some features of Slicer3 are not fully tested.
- The Slicer Training page provides a series of courses for learning how to use Slicer3. The portfolio contains self-guided presentation and sample data sets
The main slicer.org pages provide a guided tour to the application, training materials, and the development community. New users should start there because we try to keep the pages organized and up to date.
Our Philosophy
Slicer is a community platform created for the purpose of subject specific image analysis and visualization.
- Multi-modality imaging including, MRI, CT, US, nuclear medicine, and microscopy
- Multi organ from head to toe
- Bidirectional interface for devices
- Expandable and interfaced to multiple toolkits
There is no restriction on use, but permissions are responsibility of users. For details on the license see here
Slicer Highlights
- Slicer v3.6 - New and Improved Modules
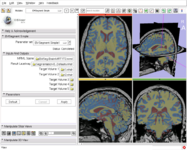
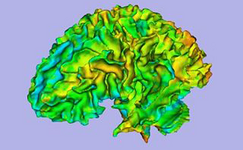
The Interactive Editor can be used to create and edit label maps for quantitative analysis and surface model generation (Steve Pieper)
Volume Rendering (Yanling Liu, Julien Finet, Lisa Avila)
Colors (Nicole Aucoin)
Measurements (rulers and angles) (Nicole Aucoin)
Fiducials (Nicole Aucoin)
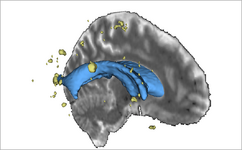
EM Segmenter (Kilian Pohl)
Fast Marching segmentation (Andriy Fedorov)
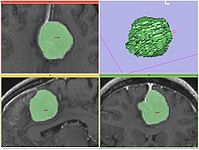
Robust Statistical Segmentation (Yi Gao)
NEW: Multiresolution Affine registration (Casey Goodlett)
Improved: Expert Automated Registration (Casey Goodlett)
NEW: Fiducial Registration (Casey Goodlett)
NEW: BRAINSFit Registration (Hans Johnson)
NEW: BRAINSDemonWarp (Hans Johnson)
NEW: BRAINSResample (Hans J. Johnson)
NEW: BRAINSROIAuto (Hans J. Johnson)
NEW: Plastimatch non-rigid registration (Greg Sharp)
PETCTFusion (Wendy Plesniak)
4D Image Viewer (Junichi Tokuda)
MRIBiasFieldCorrection correction of MRI intensity inhomogeneity i.e. bias field (Sylvain Jaume)
N4 Bias Field Correction (Andriy Fedorov)
Mesh Contour Segmentation (Peter Karasev)
Model Transform (Alex Yarmarkovich)
Resample Scalar/Vector/DWI Volume (Francois Budin)
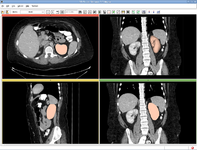
Crop Volume (Andriy Fedorov)
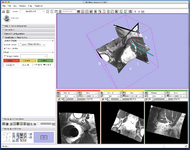
Virtual Endoscopy (Steve Pieper)
ChangeTracker (Andriy Fedorov)
OpenIGTLinkIF (Junichi Tokuda)
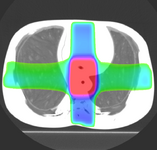
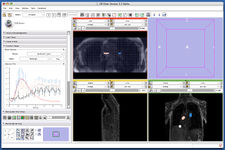
ProstateNav (Andras Lasso, Junichi Tokuda)
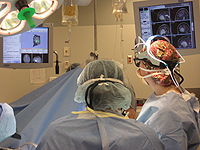
NeuroNav (Haiying Liu)
IGT tool selector (A. Wiles)
Slicer Extensions
- Slicer v3.6 - Extensions
The Plastimatch DICOM RT reader allows import and conversion of data in that format. (Greg Sharp)
The ABC Segmenter is based on ITK EM technology.
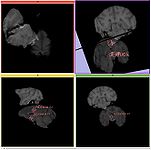
Hammer Registration can be used to build statistical brain atlases
Plastimatch non-rigid registration (Greg Sharp)
Arctic wizard (Automatic Regional Cortical ThICkness)
Lupus white matter lesions segmentation (Jeremy Bockholt, Mark Scully)
EM DTI clustering (Mahnaz Maddah)
Rician Noise Filter for noise removal in DWI data
The FourD Analysis module was designed for time series analysis (Junichi Takuda)
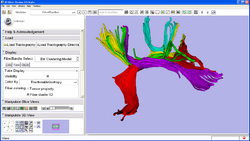
Centerline extraction using Voronoi diagrams (Luca Antiga, Daniel Haehn)
Label diameter estimation (Andriy Fedorov)
Checklist for the 3.6 release
Other Improvements, Additions & Documentation
New default LUT (Michael Halle)
Registration Portal Page: overview of avail. tools & modules (Dominik Meier)
Resampling Tool Feature Matrix (Dominik Meier)
Slicer Registration Case Library (Dominik Meier)
Registration Welcome Module gives an overview of avail. registration modules
Segmentation Welcome Module gives an overview of avail. segmentation modules
Diffusion Welcome Module gives an overview of avail. DWI processing modules
ITK upgrades provided performance improvements in all registration modules and many other modules.