Difference between revisions of "Documentation/4.4/Announcements"
(Nightly -> 4.4) |
|||
| Line 23: | Line 23: | ||
The [http://www.slicer.org/pages/Acknowledgments community] of Slicer developers is proud to announce the release of '''Slicer {{documentation/version}}'''. | The [http://www.slicer.org/pages/Acknowledgments community] of Slicer developers is proud to announce the release of '''Slicer {{documentation/version}}'''. | ||
| + | For the online documentation pages please click [[Documentation/{{documentation/version}} | here]]. | ||
* Slicer {{documentation/version}} introduces | * Slicer {{documentation/version}} introduces | ||
** An improved App Store, known as the Extension Manager, for adding plug-ins to Slicer. More than 50 plug-ins and packages of plug-ins are currently available. | ** An improved App Store, known as the Extension Manager, for adding plug-ins to Slicer. More than 50 plug-ins and packages of plug-ins are currently available. | ||
Latest revision as of 15:27, 23 March 2015
Home < Documentation < 4.4 < Announcements
|
For the latest Slicer documentation, visit the read-the-docs. |
| Summary | What is 3D Slicer | Slicer 4.4 Highlights | Slicer Training | Slicer Extensions | Other Improvements, Additions & Documentation |
Summary
The community of Slicer developers is proud to announce the release of Slicer 4.4.
For the online documentation pages please click here.
- Slicer 4.4 introduces
- An improved App Store, known as the Extension Manager, for adding plug-ins to Slicer. More than 50 plug-ins and packages of plug-ins are currently available.
- Close to 400 feature improvements and bug fixes have resulted in improved performance and stability.
- Improvements to many modules.
- Click here to download Slicer 4.4 for different platforms and find pointers to the source code, mailing lists and the bug tracker.
- Please note that Slicer continues to be a research package and is not intended for clinical use. Testing of functionality is an ongoing activity with high priority, however, some features of Slicer are not fully tested.
- The Slicer Training page provides a series of tutorials and data sets for training in the use of Slicer.
slicer.org is the portal to the application, training materials, and the development community.
What is 3D Slicer
3D Slicer is:
- A software platform for the analysis (including registration and interactive segmentation) and visualization (including volume rendering) of medical images and for research in image guided therapy.
- A free, open source software available on multiple operating systems: Linux, MacOSX and Windows
- Extensible, with powerful plug-in capabilities for adding algorithms and applications.
Features include:
- Multi organ: from head to toe.
- Support for multi-modality imaging including, MRI, CT, US, nuclear medicine, and microscopy.
- Bidirectional interface for devices.
There is no restriction on use, but Slicer is not approved for clinical use and intended for research. Permissions and compliance with applicable rules are the responsibility of the user. For details on the license see here
Slicer 4.4 Highlights
- New and Improved Modules
- Improved Transforms module with support for non-linear transforms, visualization of transforms in 2D and 3D, detailed transform properties view - click here for demo video.
- Improved integration of non rigid deformations, including grid (displacement field) and bspline transforms
- Interactive application of non rigid deformations to volume slices, models, markups
- Visualization of any transforms as glyphs, grid, or contours in 2D slice and 3D views
- Computing and applying inverse transforms, compositing any number of transforms
- Real-time update: if the transform (or any visualization parameter) is changed then the visualization is updated immediately (interactive visualization while editing the transform)
- Detailed transform information display (type of transform, basic properties, displacement at current position)
- Loading/saving of oriented bspline transforms with or without additive bulk component
- Loading/saving of oriented grid transforms
- Loading/saving of transforms in h5 file format
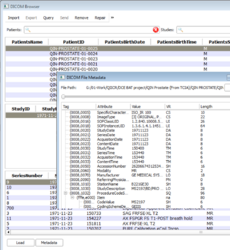
- The user interface of the DICOM module has been improved for a better usability. A viewer for meta-data (header) has been added.
- Added a way to change the table densities in three levels: compact, cozy and comfortable.
- Re-arranged the patient, study and series search box so that the search box will be on the top of tables for both horizontal and vertical cases.
- Resize tables based on contents.
- Allow users to acknowledge all DICOM loading errors with a single click
- Add DICOM meta-data (header) viewer
- Auto-examine in dicom browser added by providing two modes: Advanced and Non-advanced mode.
- Added repair tool for DICOM database
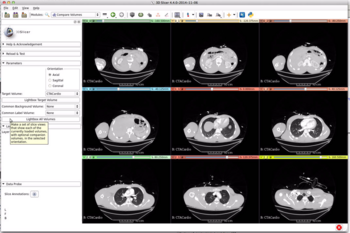
- Compare Volumes provides the ability to create an overview of one or more volumes.
- For one volume you can look at an array of equally spaced images (like LightBox mode, but wth an independent viewer per slice.
- You can have a look at one viewer per loaded volume.
- You can choose a common label or background volume for each per-volume viewer.
- The RevealCursor allows you to look in detail at the fg/bg layers.
- Improvements to the Interactive Editor
- Added a Sphere option to the PaintEffect to make it quicker to segment large anatomical regions. Both the tumor and part of the superior saggital sinus were labeled using this effect.
- Paint using pixel mode if brush size is too small
- Button effect are checked/unchecked based on the 'effect' property stored in the parameter node
The LandmarkRegistration module was custom designed for interactive registration.
New annotations can be controlled in the DataProbe module. This new feature allows to display corner annotations (with information about the volumes available in the different layers of the Slice Viewer), a color scalar bar and an interactive scaling ruler.
The Scene Views module GUI updated with better layout.
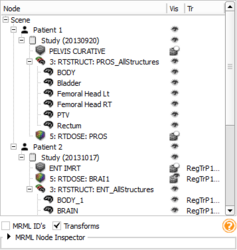
The Subject hierarchy module allows to organize and manipulate data loaded in Slicer.
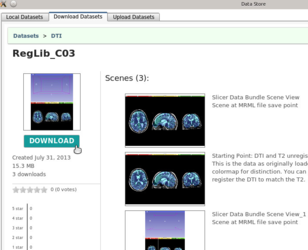
The DataStore module allows easy upload and download of MRB files from a Midas server.
- The ExtensionsWizard replaces the ModuleWizard.
- This new tool separates the concepts of extensions and modules, and allows creating an extension containing several modules, as well as adding modules to an existing extension.
- It allows to publish the extension source code to github.
- It provides an easy way to create a pull request on the extension index.
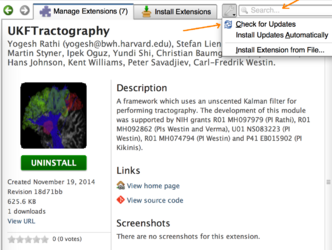
- Improved ExtensionsManager:
- Improved download to install extension dependencies
- Added search box to the ExtensionsManager
- Added mechanism to check for extensions updates either manually or automatically
- Added support for Installing extension without network connection
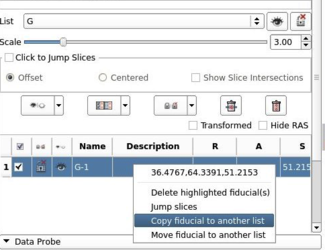
- Improved Markups module user interface.
- Added slice intersections toggle
- Added right click option to copy markups list
- Added coordinates to right click menu
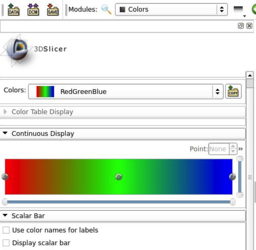
- Improved Colors module user interface.
- Support for continuous color maps
- Added an option to use color names as labels on the scalar bar
Slicer Training
The Slicer Training page provides a series of updated tutorials and data sets for training in the use of Slicer 4.4.
- New Tutorials
A hands-on session called Contributing extensions to 3D Slicer was conducted during the 3D Slicer workshop and training at the University of Iowa on Nov 18th, 2014 (Andriy Fedorov, Jean-Christophe Fillion-Robin) NEW
Slicer IGT Tutorials: using Slicer for image-guided therapy (ultrasound, CT, MRI guidance, optical and electromagnetic navigation, etc.)
Slicer Extensions
- New and Improved Extensions
Airway Segmentation to segment the airway from chest CT images. UPDATED
CardiacAgastonMeasures to auto-segment the coronary calcium in Cardiac CT scans and then calculate the Agatston score and label statistics NEW
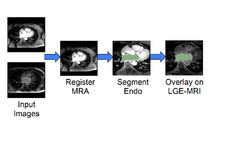
Cardiac MRI Toolkit to analyze cardiac LGE-MRI images. UPDATED
CarreraSliceInteractiveSegmenter to interactively segment in 3D UPDATED
CBC 3D (CRTC's BCC & Compression) I2M (Image-To-Mesh) Conversion for Image Guided Therapy. The extension encapsulates two CLI modules: (1) Body Centric Cubic (BCC) Mesh Generation. (2) Mesh Compression (MC). NEW
Change Tracker for quantification of the subtle changes in pathology. UPDATED
CleaverExtension extension bundles the Cleaver meshing tool as a CLI module NEW
Dice Computation to calculate the Dice Similarity Coefficient (DSC) between multiple label map images. UPDATED
CMFreg is a set of tools package for cranio-maxillofacial registration UPDATED
CornerAnnotation is a module to display annotations, time count, node elements on each panels on the Slicer user interface UPDATED
CurveMaker is a module to generate a curve based on a list of fiducial points. NEW
DTI Prep: The DWI/DTI Quality Control Processes from DICOM data to qualified DWI image. UPDATED
DTI Process: DTI processing and analysis toolkit developed at UNC and University of Utah. Tools in this toolkit include (1) dtiestim, (2) dtiprocess, (3) dtiaverage, (4) fiberprocess and (5) fiberstats. UPDATED
FastGrowCut is an editor effect providing a fast implementation of the GrowCut method that supports multi-label segmentations UPDATED
FinslerTractography implements the Finsler tractography method with HARDI data as described by J. Melonakos et al., Finsler Active Contours, IEEE Trans PAMI, 30:412-423, 2008. NEW
GelDosimetryAnalysis is a Slicelet covering the gel dosimetry analysis workflow used in commissioning new radiation techniques. UPDATED
GyroGuide determines the trajectory angle and depth of puncture path, and transmits the calculated information to the probe for real time navigation. NEW
IASEM to segmentation and process of IASEM Electron Microscopy images. UPDATED
iGyne is an open source software for MR-Guided Interstitial Gynecologic Brachytherapy. NEW
IntensitySegmenter is a simple tool that segments an image according to intensity value. It is mainly used to segment CT scans using the Hounsfield scale but the ranges of intensities and their corresponding labels can be specified in an input text file. UPDATED
LightWeightRobotIGT to manage communication between 3D Slicer and LightWeight robot. NEW
Longitudinal PET/CT to provide a user friendly Slicer interface for quantification of DICOM PET/CT image data.{{updated}
MABMIS: Multi-Atlas Based Group Segmentation NEW
Matlab Bridge to allow running Matlab functions directly in 3D Slicer. UPDATED
MultidimData: A set of modules for generic multidimensional data management in Slicer (0.2.1) NEW
NeedleFinder: NeedleFinder: fast interactive needle detection. It provides interactive tools to segment needles in MR/CT images. It has been mostly tested on MRI from gynelogical brachytherapy cases. NEW
PBNRR: This extension encapsulates a CLI module for the Physics-Based Non-Rigid Registration (PBNRR) method. The PBNRR compensates for the brain shift during the Image-Guided Neurosurgery (IGNS). NEW
PercutaneousApproachAnalysis: The Percutaneous Approach Analysis is used to calculate and visualize the accessibility of liver tumor with a percutaneous approach. NEW
PerkTutor for training in image-guided needle interventions. UPDATED
PETDICOM: The PET DICOM Extension provides tools to import PET Standardized Uptake Value images from DICOM into Slicer. NEW
Pk Modeling to calculate quantitative parameters from Dynamic Contrast Enhanced DCE-MRI images. UPDATED
Port Placement to assists in the planning of surgical port placement in laparoscopic procedures. UPDATED
PyDevRemoteDebug: This extension allows remote visual debugging of Python scripts using PyDev (http://pydev.org/) NEW
Reporting to create image annotations/markup that are stored in a structured form. UPDATED
ResectionPlanner: Modules for surgical resection planning. NEW
Scoliosis: Extensions pertaining to scoliosis analysis UPDATED
ShapePopulationViewer: Visualize and interact with multiple surfaces at the same time to easily compare them UPDATED
http://slicer.vmtk.org/ SlicerExtension-VMTK]: The Vascular Modeling Toolkit as a 3D Slicer4 extension. NEW
SlicerIGT to use all the advanced features of 3D Slicer for real-time navigation. UPDATED
SlicerRT is a tool for powerful radiotherapy research. UPDATED
TCIABrowser: A Module to connect to TCIA archive, browse the collections, patients and studies and download DICOM files to 3D Slicer. NEW
Tracker Stabilizer to output a filtered transform node based on an tracker input (transform node). UPDATED
UKF Tractography a framework which uses an unscented Kalman filter for performing tractography. UPDATED
VolumeClip: Clip volumes with surface models and ROI boxes NEW
Window/Level Effect to adjust window/level for volumes using mouse and/or region of interest. UPDATED
XNAT Slicer Secure GUI-based IO with any XNAT server. UPDATED
Extensions removed
- DataStore: The module is now bundled in the regular Slicer distribution.
- PathPlanner and VisualLine: Module PathXplorer available in SlicerIGT extension provides similar functionality.
- PlusRemote: Moved PlusRemote module under SlicerIGT extension. Because PlusRemote depends on SlicerIGT, and is always used together with the other SlicerIGT modules.
- TransformVisualizer: The feature has been moved to the Slicer core. See Transforms module.
Extensions renamed
- VMTKSlicerExtension -> SlicerExtension-VMTK
- KSlice -> CarreraSliceInteractiveSegmenter
Other Improvements, Additions & Documentation
Optimization
- Improved performance of the rendering pipeline by optimizing observations management
- Reduced the memory footprint by fixing memory leaks
- Reduced the size of the installer
- Faster loading of the image stack (png, jpg, bmp, tiff...)
Rendering / Visualization
- Added Multi-sampling option
CLI / SlicerExecutionModel
- Improved CLI AutoRun by ensuring that the slice view is not reset when inputs are updated.
- CLI input and output files are not removed when in developer mode
- Reduced the chance of crashes when CLI returns result images
Python scripting
- Added support for real Qt resources in Python (see r23290 for details).
- Improved VTK event support by adding a way to specify the CallData type (see here for details).
- Bundled pydicom python module into the Slicer package
- Added helper methods to
ScriptedLoadableModuleLogicfor managing parameter nodes
- SelfTest: Added the
clickAndDrag()method to the scripted module construct. It allows to send synthetic mouse events to the widget specified (qMRMLSliceWidget or qMRMLThreeDView)
- Improvements to the
slicer.utilpython module.- Added
modulePath(moduleName)method - Added
resetThreeDViews/resetSliceViews - Added
VTKObservationMixin - Added
getFirstNodeByClassAndName() - Added
NodeModifycontext manager: It allows to easily disable modified event associated with a node, and automatically re-enable them and invoking them if it applies.
- Added
- Markups: Added an event called PointEndInteractionEvent for marking the end of a fiducial interaction
- Reference information for how to write VTK filters and algorithms in python:
- The vtkPythonAlgorithm is great
- A VTK pipeline primer (part 1): Explains how VTK’s pipeline works in more detail:
vtkInformation,vtkInformationVectorandVTKPythonAlgorithmBase - A VTK pipeline primer (part 2): Dissecting the execution path to understand the inner-workings of algorithms:
RequestInformation - A VTK pipeline primer (part 3): Explains how
RequestUpdateExtentandRequestDatawork.
Other
- Help / Report a bug: Application error/warning/debug log messages are now saved to file. Added option to copy/paste log file contents of recent Slicer sessions to bug reports.
For Developers
Under the hood
- VolumeLogic:
- Added method ResampleVolumeToReferenceVolume
- Added method CloneVolumeWithoutImageData
- Added support for user-defined stereo-viewing options
- Build-system
- Improved support for Visual Studio 2013
- Refactored management of external project launcher settings. See r23724
- Added option
Slicer_ITKV3_COMPATIBILITY. This option enabled by default will allow (if disabled) to build Slicer with ITKv3 compatibility later disabled andITK_USE_64BITS_IDSenabled.
- SlicerExecutionModel: Added ParameterSerializer support.
- Improved Toolkits
Moved from OpenIGTLink 66e272d to 849b434 (53 commits)
Looking at the Code Changes
From a git checkout you can easily see the all the commits since the time of the 4.4.0 release:
git log v4.3.0..HEAD
To see a summary of your own commits, you could use something like:
git log v4.3.0..HEAD --oneline --author=pieper
see the git log man page for more options.
Commit stats and full changelog