Difference between revisions of "Registration:Categories"
From Slicer Wiki
| (21 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
= <small><small>looking for a set of features:</small></small> 3DSlicer Registration Feature/Decision Matrix [[Image:Registration_DecisionMatrix.png|70px]] = | = <small><small>looking for a set of features:</small></small> 3DSlicer Registration Feature/Decision Matrix [[Image:Registration_DecisionMatrix.png|70px]] = | ||
| − | *the matrix below shows which modules support which feature | + | *the matrix below shows which modules support which feature, intended to help you if looking for a method with a particular combination of features. |
*if you would like to see additional criteria or a new module is not yet listed, please let us know: mailto:slicer-users at bwh.harvard.edu | *if you would like to see additional criteria or a new module is not yet listed, please let us know: mailto:slicer-users at bwh.harvard.edu | ||
| − | [[Image:Registration_DecisionMatrix.png|left|850px|3DSlicer Registration Feature/Decision Matrix]] | + | *see here for an [http://labelpage.halle.us/static/reg/index.html '''interactive method selection tool'''] |
| + | [[Image:Registration_DecisionMatrix.png|left|850px|3DSlicer Registration Feature/Decision Matrix]] | ||
= <small><small>looking for</small></small> Speed [[Image:Registration_Speed_icon.png|70px]] = | = <small><small>looking for</small></small> Speed [[Image:Registration_Speed_icon.png|70px]] = | ||
| − | *'''Manual'''/interactive alignment can be done via the [[Modules:Transforms-Documentation-3. | + | *'''Manual'''/interactive alignment can be done via the [[Modules:Transforms-Documentation-3.6|'''Transforms''' ]] module, e.g. for initial alignment. See [[Slicer3.4:Training#Slicer_3.4_Tutorials| here for a tutorial and example dataset on Manual Registration]] |
| − | *'''Affine:''' The [[Modules:AffineRegistration-Documentation-3. | + | *'''Affine:''' The [[Modules:AffineRegistration-Documentation-3.6|'''Fast Affine Registration''']] module performs automated affine registration. |
| − | *'''Bspline:''' The [[Modules:DeformableB-SplineRegistration-Documentation-3. | + | *'''Bspline:''' The [[Modules:DeformableB-SplineRegistration-Documentation-3.6|'''Fast Nonrigid BSpline Registration''']] module performs non-rigid automated image registration. With default settings results within <1 minute are common. |
| + | *The [[Modules:ACPCTransform-Documentation-3.6|'''AC-PC Transform''']] module is used to orient '''brain''' images along predefined anatomical landmarks: (manually defined) fiducials for the inter-hemispheral midline. Once landmarks are defined, realignment is obtained instantly. | ||
= <small><small>looking for</small></small> Robustness and/or Precision [[Image:Registration_Precision_icon.png| 70px]] = | = <small><small>looking for</small></small> Robustness and/or Precision [[Image:Registration_Precision_icon.png| 70px]] = | ||
| − | *The [[Modules:RegisterImages-Documentation-3. | + | *The [[Modules:RegisterImages-Documentation-3.6|'''Expert Automated Registration''']] module performs automated image registration, rigid to affine, based on image intensity similarities. It allows to focus the registration on a region of interest |
| − | *The [[Modules:RegisterImagesMultiRes-Documentation-3.6|''' | + | *The [[Modules:RegisterImagesMultiRes-Documentation-3.6|'''Robust Multiresolution Affine Registration''']] module performs robust automated affine image registration employing a multi-resolution scheme. |
| − | *Plastimatch | + | *The [[Modules:Plastimatch|'''Plastimatch''']] module performs automated registration of images from rigid to affine to non-rigid. As a unique feature it provides non-rigid deformation from fiducials, which can be used to "edit/repair" a registration. |
| + | *The [[Modules:ACPCTransform-Documentation-3.6|'''AC-PC Transform''']] module is used to orient '''brain''' images along the anterior- and posterior commissure and midline. Within the accuracy of the manually chosen anatomical landmarks this alignment tends to be very robust. | ||
= <small><small>choose by</small></small> DOF [[Image:Registration_HLogo_DOF.png| 70px]] = | = <small><small>choose by</small></small> DOF [[Image:Registration_HLogo_DOF.png| 70px]] = | ||
*'''rigid 6 DOF:''' | *'''rigid 6 DOF:''' | ||
| − | **[[Modules:RegisterImages-Documentation-3. | + | **[[Modules:RegisterImages-Documentation-3.6|'''Expert Automated Registration''']]. Set DOF to ''PipelineRigid''. |
*'''similarity 9 DOF:''' | *'''similarity 9 DOF:''' | ||
| + | **The [[Modules:BRAINSFit| '''BAINSfit''']] module includes similarity transform; type ''ScaleVersor3D'' in the transform type field; provides masking support. | ||
| + | **The [[Modules:PythonSurfaceICPRegistration-Documentation-3.6|'''Surface Registration''' ]] module supports automated registration of surfaces with rigid or similarity transforms. | ||
| + | **The [[Modules:TransformFromFiducials-Documentation-3.6|'''Fiducial Alignment''']] module supports a similarity transform. Two sets of fiducials (fiducial lists) are required, forming matching pairs to be aligned. See ''Fiducials'' module. | ||
*'''affine 12 DOF:''' | *'''affine 12 DOF:''' | ||
| − | **[[Modules:AffineRegistration-Documentation-3. | + | **[[Modules:AffineRegistration-Documentation-3.6|'''Fast Affine Registration''']] |
| − | **The [[Modules:RegisterImagesMultiRes-Documentation-3.6|''' | + | **The [[Modules:RegisterImagesMultiRes-Documentation-3.6|'''Robust Multiresolution Affine Registration''']] module performs robust affine (12 DOF) image registration employing a multi-resolution scheme. |
| − | **[[Modules:RegisterImages-Documentation-3. | + | **[[Modules:RegisterImages-Documentation-3.6|'''Expert Automated Registration''']]. Set DOF to ''PipelineAffine''. |
*'''non rigid 27- 100s DOF:''' | *'''non rigid 27- 100s DOF:''' | ||
| − | ** The [[Modules:DeformableB-SplineRegistration-Documentation-3. | + | ** The [[Modules:DeformableB-SplineRegistration-Documentation-3.6|'''Fast Nonrigid BSpline Registration''']] module performs non-rigid automated image registration. |
| + | **The [[Modules:BRAINSFit| '''BAINSfit''']] module includes a registration based on a Bspline transform. Initially designed for but not limited to '''brain''' images. Also includes many options such as masking support. | ||
*'''non rigid (fluid) >100 DOF''' | *'''non rigid (fluid) >100 DOF''' | ||
| − | **The [[Modules: | + | **The [[Modules:BRAINSDemonWarp|'''BRAINSDemonWarp''' ]] module performs automated registration of brain MRI based on an optic flow mechanism. Deformations here are significantly more "fluid" (i.e. have more DOF and are less constrained) than for the nonrigid BSpline method. |
| − | **The [ | + | **The [[Modules:Plastimatch|'''Plastimatch''']] module performs automated registration of images from rigid to affine to non-rigid. As a unique feature it provides non-rigid deformation from fiducials, which can be used to "edit/repair" a registration. |
| − | + | **The [[Modules:HammerRegistration| '''HAMMER''']] Module performs elastic (non-rigid) alignment of '''brain''' images of different individuals based on tissue class segmentation and intensity (experimental stage). | |
| − | |||
| − | |||
| − | |||
| − | = <small><small>choose by</small></small>Datatype [[Image:Registration_HLogo_Datatype.png| 70px]] = | + | = <small><small>choose by</small></small> Datatype [[Image:Registration_HLogo_Datatype.png| 70px]] = |
| − | *'''images, same modality:''' The [[Modules:RegisterImages-Documentation-3. | + | *'''images, same modality:''' The [[Modules:RegisterImages-Documentation-3.6|'''Expert Automated Registration''']] module performs automated image registration, rigid to affine, based on image intensity similarities. It allows to focus the registration on a region of interest |
| − | *'''images, different modality:''' The [[Modules:RegisterImages-Documentation-3. | + | *'''images, different modality:''' The [[Modules:RegisterImages-Documentation-3.6|'''Expert Automated Registration''']] module performs automated image registration, rigid to affine, based on image intensity similarities. Select Mutual Information as cost function. |
| − | *'''surfaces:''' The [[Modules:PythonSurfaceICPRegistration-Documentation-3. | + | *'''surfaces:''' The [[Modules:PythonSurfaceICPRegistration-Documentation-3.6|'''Surface Registration''' ]] module performs automated registration of surfaces (not images). This is useful if image data directly is unreliable, but surfaces can be produced from segmentations that provide good information about desired alignment. |
| − | *'''fiducials:''' The [[Modules: | + | *'''fiducials:''' The [[Modules:TransformFromFiducials-Documentation-3.6|'''Fiducial Alignment''']] module can align images based on pairs of manually selected fiducial points (rigid and affine). Two sets of fiducials (fiducial lists) are required, forming matching pairs to be aligned. Use the [[Modules:Fiducials-Documentation-3.6|'''Fiducials''' module]] to create fiducial lists. The [[Modules:ACPCTransform-Documentation-3.6|'''AC-PC Transform''']] module is also a special form of fiducial registration, used to orient '''brain''' images along the anterior- and posterior commissure and midline. |
| + | |||
| + | = <small><small>looking for</small></small> Brain Image Registration [[Image:Registration_HLogo_Brain.png| 70px]] = | ||
| + | *The [[Modules:ACPCTransform-Documentation-3.6|'''AC-PC Transform''']] module is used to orient '''brain''' images along predefined anatomical landmarks: (manually defined) fiducials for the inter-hemispheral midline, anterior- and posterior commissure are used to align an image such that these landmarks become vertical and horizontal, respectively. | ||
| + | **The [[Modules:BRAINSFit| '''BAINSfit''']] module includes a registration based on a Bspline transform. Initially designed for but not limited to '''brain''' images. Also includes many options such as masking support. | ||
| + | *The [[Modules:BRAINSDemonWarp|'''BRAINSDemonWarp''' ]] module performs automated registration of brain MRI based on an optic flow mechanism. Deformations here are significantly more "fluid" (i.e. have more DOF and are less constrained) than for the nonrigid BSpline method. | ||
| + | *The [[Modules:HammerRegistration| '''HAMMER''']] Module performs elastic (non-rigid) alignment of '''brain''' images of different individuals based on tissue class segmentation and intensity (experimental stage). | ||
| + | |||
| + | |||
| + | = <small><small>looking for</small></small> Tools to Resample Data [[Image:Registration_Resample_icon.png| 70px]] = | ||
| + | *see the [[Registration:Resampling|'''Overview of Resampling Tools''']] for all available resampling methods, including tools to resample in place (e.g. change resolution or voxel anisotropy etc.) | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
= <small><small>looking for</small></small> Tools for Preparing Data for Registration [[Image:Registration_Masking_icon.png| 70px]] = | = <small><small>looking for</small></small> Tools for Preparing Data for Registration [[Image:Registration_Masking_icon.png| 70px]] = | ||
*Intensity Normalization & Filtering | *Intensity Normalization & Filtering | ||
:Intensity corrections are often the first processing step of choice. MRI bias field inhomogeneities can adversely affect registration accuracy and stability, as can large differences in the intensity ranges between the two images, or large amounts of noise | :Intensity corrections are often the first processing step of choice. MRI bias field inhomogeneities can adversely affect registration accuracy and stability, as can large differences in the intensity ranges between the two images, or large amounts of noise | ||
| − | :*[[Modules:MRIBiasFieldCorrection-Documentation-3. | + | :*[[Modules:MRIBiasFieldCorrection-Documentation-3.6|Bias Field Correction to remove intensity drift across the image (e.g from variable coil sensitivity)]] |
:*[[Modules:HistogramMatching-Documentation-3.6|Histogram Equalization for matching intensity ranges]] | :*[[Modules:HistogramMatching-Documentation-3.6|Histogram Equalization for matching intensity ranges]] | ||
:*[[Modules:MedianFilter-Documentation-3.6|Median Filtering to remove speckle noise without smoothing edges]] | :*[[Modules:MedianFilter-Documentation-3.6|Median Filtering to remove speckle noise without smoothing edges]] | ||
| − | :*[[Modules:GaussianBlur-Documentation-3. | + | :*[[Modules:GaussianBlur-Documentation-3.6|Gaussian Blur]] filter to remove noise and smooth edges |
:*[[Documentation-3.6#Filtering|other filters see here]] | :*[[Documentation-3.6#Filtering|other filters see here]] | ||
*Masking | *Masking | ||
:Masks are an important component of many registration tasks. They allow to focus the algorithm on the region of interest (ROI) that is to be registered and prevent it from being distracted by image content outside this ROI. | :Masks are an important component of many registration tasks. They allow to focus the algorithm on the region of interest (ROI) that is to be registered and prevent it from being distracted by image content outside this ROI. | ||
| − | :* [http://www.na-mic.org/Wiki/index.php/2009_Summer_Project_Week_Skull_Stripping Skull Stripping] | + | :* [http://www.na-mic.org/Wiki/index.php/2009_Summer_Project_Week_Skull_Stripping Skull Stripping] extension module automatically builds a mask of the brain from an input MRI image (T1w is best). This is an extension module and needs to be installed via the Extension manager. |
| − | :*[[Modules:Editor-Documentation-3. | + | :*[[Modules:Editor-Documentation-3.6|Thresholding within the Interactive Editor]] or via the [[Modules:ThresholdImage-Documentation-3.6|Threshold module]]. |
:*[[Modules:OtsuThresholdSegmentation-Documentation-3.6|Otsu Threshold Segmentation]] for automated segmentation of main image object from background | :*[[Modules:OtsuThresholdSegmentation-Documentation-3.6|Otsu Threshold Segmentation]] for automated segmentation of main image object from background | ||
:*[[Modules:FastMarchingSegmentation-Documentation-3.6|Fast Marching segmentation]] for interactive fiducial-based segmentation of small local regions. | :*[[Modules:FastMarchingSegmentation-Documentation-3.6|Fast Marching segmentation]] for interactive fiducial-based segmentation of small local regions. | ||
:*[[Modules:MaskImage-Documentation-3.6|Mask image module]] to apply a mask and remove image content outside (if mask is not directly supported by the chosen registration method) | :*[[Modules:MaskImage-Documentation-3.6|Mask image module]] to apply a mask and remove image content outside (if mask is not directly supported by the chosen registration method) | ||
| − | :*[[Modules:ExtractSubvolumeROI-Documentation-3. | + | :*[[Modules:ExtractSubvolumeROI-Documentation-3.6|Crop/SubvolumeExtraction module]] to extract a box region from the image. |
:*[[Modules:ROIModule-Documentation-3.6|ROI module]] to quickly define a box region of interest that is supported as mask by [[Modules:RegisterImagesMultiRes-Documentation-3.6|Robust Multires Registration]], for example. | :*[[Modules:ROIModule-Documentation-3.6|ROI module]] to quickly define a box region of interest that is supported as mask by [[Modules:RegisterImagesMultiRes-Documentation-3.6|Robust Multires Registration]], for example. | ||
| − | |||
| − | |||
Latest revision as of 13:07, 30 June 2010
Home < Registration:CategoriesBack to registration portal page
Slicer Registration Case Library: Examples & Tutorials
Contents
- 1 looking for a set of features: 3DSlicer Registration Feature/Decision Matrix
- 2 looking for Speed
- 3 looking for Robustness and/or Precision
- 4 choose by DOF
- 5 choose by Datatype
- 6 looking for Brain Image Registration
- 7 looking for Tools to Resample Data
- 8 looking for Tools for Preparing Data for Registration
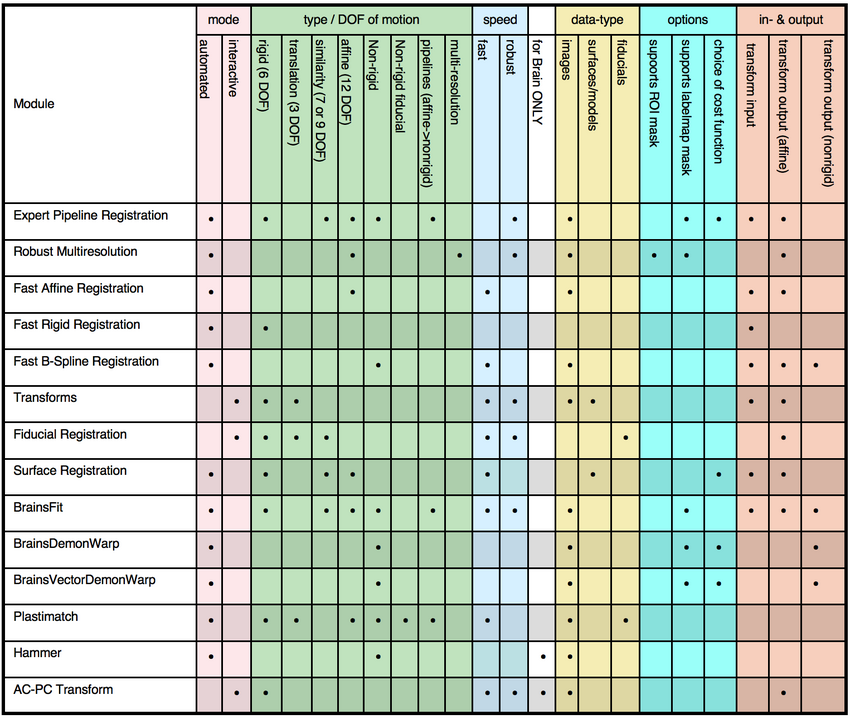
looking for a set of features: 3DSlicer Registration Feature/Decision Matrix 
- the matrix below shows which modules support which feature, intended to help you if looking for a method with a particular combination of features.
- if you would like to see additional criteria or a new module is not yet listed, please let us know: mailto:slicer-users at bwh.harvard.edu
- see here for an interactive method selection tool
looking for Speed 
- Manual/interactive alignment can be done via the Transforms module, e.g. for initial alignment. See here for a tutorial and example dataset on Manual Registration
- Affine: The Fast Affine Registration module performs automated affine registration.
- Bspline: The Fast Nonrigid BSpline Registration module performs non-rigid automated image registration. With default settings results within <1 minute are common.
- The AC-PC Transform module is used to orient brain images along predefined anatomical landmarks: (manually defined) fiducials for the inter-hemispheral midline. Once landmarks are defined, realignment is obtained instantly.
looking for Robustness and/or Precision 
- The Expert Automated Registration module performs automated image registration, rigid to affine, based on image intensity similarities. It allows to focus the registration on a region of interest
- The Robust Multiresolution Affine Registration module performs robust automated affine image registration employing a multi-resolution scheme.
- The Plastimatch module performs automated registration of images from rigid to affine to non-rigid. As a unique feature it provides non-rigid deformation from fiducials, which can be used to "edit/repair" a registration.
- The AC-PC Transform module is used to orient brain images along the anterior- and posterior commissure and midline. Within the accuracy of the manually chosen anatomical landmarks this alignment tends to be very robust.
choose by DOF 
- rigid 6 DOF:
- Expert Automated Registration. Set DOF to PipelineRigid.
- similarity 9 DOF:
- The BAINSfit module includes similarity transform; type ScaleVersor3D in the transform type field; provides masking support.
- The Surface Registration module supports automated registration of surfaces with rigid or similarity transforms.
- The Fiducial Alignment module supports a similarity transform. Two sets of fiducials (fiducial lists) are required, forming matching pairs to be aligned. See Fiducials module.
- affine 12 DOF:
- Fast Affine Registration
- The Robust Multiresolution Affine Registration module performs robust affine (12 DOF) image registration employing a multi-resolution scheme.
- Expert Automated Registration. Set DOF to PipelineAffine.
- non rigid 27- 100s DOF:
- The Fast Nonrigid BSpline Registration module performs non-rigid automated image registration.
- The BAINSfit module includes a registration based on a Bspline transform. Initially designed for but not limited to brain images. Also includes many options such as masking support.
- non rigid (fluid) >100 DOF
- The BRAINSDemonWarp module performs automated registration of brain MRI based on an optic flow mechanism. Deformations here are significantly more "fluid" (i.e. have more DOF and are less constrained) than for the nonrigid BSpline method.
- The Plastimatch module performs automated registration of images from rigid to affine to non-rigid. As a unique feature it provides non-rigid deformation from fiducials, which can be used to "edit/repair" a registration.
- The HAMMER Module performs elastic (non-rigid) alignment of brain images of different individuals based on tissue class segmentation and intensity (experimental stage).
choose by Datatype 
- images, same modality: The Expert Automated Registration module performs automated image registration, rigid to affine, based on image intensity similarities. It allows to focus the registration on a region of interest
- images, different modality: The Expert Automated Registration module performs automated image registration, rigid to affine, based on image intensity similarities. Select Mutual Information as cost function.
- surfaces: The Surface Registration module performs automated registration of surfaces (not images). This is useful if image data directly is unreliable, but surfaces can be produced from segmentations that provide good information about desired alignment.
- fiducials: The Fiducial Alignment module can align images based on pairs of manually selected fiducial points (rigid and affine). Two sets of fiducials (fiducial lists) are required, forming matching pairs to be aligned. Use the Fiducials module to create fiducial lists. The AC-PC Transform module is also a special form of fiducial registration, used to orient brain images along the anterior- and posterior commissure and midline.
looking for Brain Image Registration 
- The AC-PC Transform module is used to orient brain images along predefined anatomical landmarks: (manually defined) fiducials for the inter-hemispheral midline, anterior- and posterior commissure are used to align an image such that these landmarks become vertical and horizontal, respectively.
- The BAINSfit module includes a registration based on a Bspline transform. Initially designed for but not limited to brain images. Also includes many options such as masking support.
- The BRAINSDemonWarp module performs automated registration of brain MRI based on an optic flow mechanism. Deformations here are significantly more "fluid" (i.e. have more DOF and are less constrained) than for the nonrigid BSpline method.
- The HAMMER Module performs elastic (non-rigid) alignment of brain images of different individuals based on tissue class segmentation and intensity (experimental stage).
looking for Tools to Resample Data 
- see the Overview of Resampling Tools for all available resampling methods, including tools to resample in place (e.g. change resolution or voxel anisotropy etc.)
looking for Tools for Preparing Data for Registration 
- Intensity Normalization & Filtering
- Intensity corrections are often the first processing step of choice. MRI bias field inhomogeneities can adversely affect registration accuracy and stability, as can large differences in the intensity ranges between the two images, or large amounts of noise
- Masking
- Masks are an important component of many registration tasks. They allow to focus the algorithm on the region of interest (ROI) that is to be registered and prevent it from being distracted by image content outside this ROI.
- Skull Stripping extension module automatically builds a mask of the brain from an input MRI image (T1w is best). This is an extension module and needs to be installed via the Extension manager.
- Thresholding within the Interactive Editor or via the Threshold module.
- Otsu Threshold Segmentation for automated segmentation of main image object from background
- Fast Marching segmentation for interactive fiducial-based segmentation of small local regions.
- Mask image module to apply a mask and remove image content outside (if mask is not directly supported by the chosen registration method)
- Crop/SubvolumeExtraction module to extract a box region from the image.
- ROI module to quickly define a box region of interest that is supported as mask by Robust Multires Registration, for example.