Difference between revisions of "Announcements:Slicer3.2"
m (Text replacement - "slicerWiki/index.php" to "wiki") |
|||
| (106 intermediate revisions by 11 users not shown) | |||
| Line 1: | Line 1: | ||
| − | =Slicer 3.2 | + | =Introduction= |
| + | |||
| + | '''Links to Latest Slicer 3.2 Binaries''' | ||
| + | |||
| + | 64-bit Linux: | ||
| + | http://www.na-mic.org/Slicer/Download/Release/linux-x86_64/Slicer3-3.2.2008-08-08-linux-x86_64.tar.gz | ||
| + | |||
| + | 32-bit Linux: | ||
| + | http://www.na-mic.org/Slicer/Download/Release/linux-x86/Slicer3-3.2.2008-08-08-linux-x86.tar.gz | ||
| + | |||
| + | Darwin PPC: | ||
| + | http://www.na-mic.org/Slicer/Download/Release/darwin-ppc/Slicer3-3.2.2008-08-08-darwin-ppc.tar.gz | ||
| + | |||
| + | Darwin x86: | ||
| + | http://www.na-mic.org/Slicer/Download/Release/darwin-x86/Slicer3-3.2.2008-08-08-darwin-x86.tar.gz | ||
| + | |||
| + | Windows: | ||
| + | http://www.na-mic.org/Slicer/Download/Release/win32/Slicer3-3.2.2008-08-08-win32.exe | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | The [[Announcments-3.2-Team|community]] of Slicer developers is proud to announce the release of Slicer 3.2. This effort is the culmination of hundreds of person years and tens of millions of dollars of effort [http://www.slicer.org/wiki/Announcements:Slicer3.2#Slicer_in_Numbers]. Slicer leverages state of the art open-source toolkits for [http://www.vtk.org visualization], [http://www.itk.org medical image analysis], [http://www.cmake.org software process], and other tools for processing and accessing data (for more information see the description of the [http://wiki.na-mic.org/Wiki/index.php/NA-MIC-Kit NA-MIC Kit]). Slicer offers these capabilities as part of the open-source framework known as the NA-MIC Kit, which facilitates on-going research in biomedical computing, supports commercialization through NA-MIC Kit components, and provides a spectrum of capabilities suitable for researchers with varying levels of computer skills. | ||
| + | |||
{| border="00" cell padding="5" cellspacing="0" | {| border="00" cell padding="5" cellspacing="0" | ||
|- | |- | ||
| rowspan="2"| | | rowspan="2"| | ||
| − | + | <big> '''Slicer 3.2'''</big> is a general purpose biomedical computing application with extensive built-in visualization and analysis capabilities, accessible through an easy to use graphical interface. For advanced users, Slicer may be extended at run-time with user-defined plug-in modules. Release candidates for this application will be available the first week of June 2008. This new release contains hundreds of changes to the software. Highlights include: | |
| − | * | + | |
| − | * | + | * Improved multi-platform support and availability of pre-compiled binaries for Windows, Mac OS X, and Linux |
| − | * | + | * New ways to display, interact and record complex visualizations of cross-sectional and derived data |
| − | + | * Advanced data fusion and registration capabilities | |
| − | + | * Segmentation tools | |
| − | | style="background: # | + | * Support for complex data such as DTI |
| + | * Interfaces to informatics frameworks | ||
| + | |||
| + | Click here to [http://www.slicer.org/pages/Downloads download] different versions of Slicer3 and find pointers to the source code, mailing lists and bug tracker. Click [[Announcements:Slicer3.2:FAQ|here]] for a FAQ. Please note that Slicer continues to be a research package and is not intended for clinical use. Testing of functionality is an ongoing activity with high priority, however, some features of Slicer3 are not fully tested. | ||
| + | |||
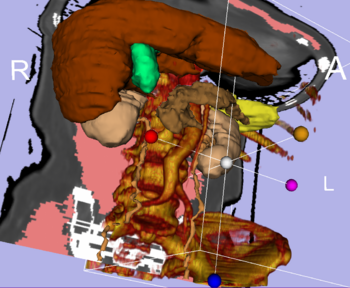
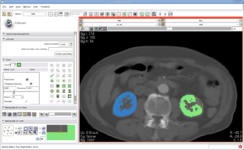
| + | | style="background: #eeeeee" align="center"| [[Modules:VolumeRendering-Documentation|Integrated Volume Rendering]]: <br>View of the [http://www.slicer.org/pages/Special:PubDB_View?dspaceid=1266 abdominal atlas]<br>Bone and large vessels are volume rendered. | ||
|- | |- | ||
| style="background: #ebeced"|[[Image:VolRend2.png|center|350px|Example of Volume Rendering]] | | style="background: #ebeced"|[[Image:VolRend2.png|center|350px|Example of Volume Rendering]] | ||
|} | |} | ||
| − | =New and | + | =Highlights= |
| + | <gallery caption="Slicer v3.2 - New and Improved Feature Highlights" widths="250px" heights="150px" perrow="3"> | ||
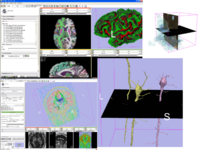
| + | Image:ComplexVis.png|Complex Visualization Capabilities: Combining cross-sections and 3D surface models tumor, brain morphology, MR angiogram, fMRI and DTI) | ||
| + | Image:VolRend.png|Volume Rendering: [[Modules:VolumeRendering-Documentation|Fully integrated volume rendering]] with cropping for easy exploration of volumetric data | ||
| + | Image:ImplicitPlane.png|Implicit Slice Widget: An interactive tool for specifying oblique views (part of the [http://www.vtk.org/Wiki/VTK_Widget_Examples VTK widget family]). | ||
| + | Image:LightBox.png|[[Slicer3:Tools_for_Radiologists|The Lightbox]]: Functionality for displaying cross-sectional data in columns and rows. | ||
| + | Image:Scenesnapshot.png| [[Modules:MainApplicationGUI-Documentation#Scene_Snapshots|Scenesnapshots]] allow to capture and recover all parameters of a scene. | ||
| + | Image:Editor-v3-2.png|[[Modules:Editor-Documentation|Interactive Editor:]] This new module allows interactive segmentation with robust 2D and 3D algorithms | ||
| + | Image:IO.png|IO: [[Documentation|Improved IO capabilities]] include support for DICOM, NRRD, NIFTI, Tiff, JPG, Freesurfer, FITS and a number of other formats | ||
| + | Image:DTI glyphs.png|Glyphs on corticospinal tract:<br>[[Slicer3:DTMRI|New Diffusion Imaging infrastructure]] includes, dicom import, gradient editor, visualiztion | ||
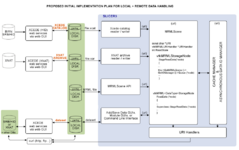
| + | Image:DataLoadingStartPlan.png|[[Modules:MainApplicationGUI-Documentation#View-.3ECache_.26_RemoteIO_Manager:|Remote Data Handling]] allows uploads and downloads from image informatics frameworks such as BIRN, and XNAT | ||
| + | Image:Slicer_IGTL_PartialImage.png|[http://wiki.na-mic.org/Wiki/index.php/IGT:ToolKit IGT Toolkit] to enable research in Image Guided Therapies | ||
| + | Image:DataFusion.png|Result of combining affine and b-Spline registration: Robust and fast (parallelized) registration algorithms. | ||
| + | Image:EMSVisualizeTutorialResultsData.png|[[Modules:EM-Segmenter-Documentation-3.2|EM Segmenter:]] A configurable image segmentation tool that uses intensity distributions along with atlas information | ||
| + | Image:CMakeLogoMediumResolution.png|[http://wiki.na-mic.org/Wiki/index.php/NAMIC_Wiki:Software_Process Quality Software Process] The NA-MIC Kit employs a test-driven software development process. Slicer3 also has a [http://wiki.na-mic.org/Wiki/index.php/NAMIC_Wiki:Software_Process#The_Slicer_Build_Process new build process]. | ||
| + | Image:LoadableModule.png|[[Slicer3:Loadable_Modules|Loadable Modules]] allows developers to provide modules to users outside of the build process, resulting in a smaller installed footprint and greater flexibility to end-users. | ||
| + | </gallery> | ||
| − | + | =Slicer in Numbers= | |
| − | + | The numbers in this table represent the components of the [http://wiki.na-mic.org/Wiki/index.php/NA-MIC-Kit NA-MIC kit]. Slicer 3 is based on the NA-MIC kit. | |
| − | + | <BR>Source: http://www.ohloh.org<br> <small> Captured on July 31 2008. The numbers in the column entitled "lines of code" are hard numbers. The other two columns are estimates based on some assumptions. Please see the [http://www.ohloh.org Ohloh] website for an explanation of how the numbers were computed. </small> | |
| − | + | {|border="1" cellpadding="1" | |
| − | + | | style="width:20%"|'''Package''' | |
| − | + | | style="width:20%"|'''Approx. # Developers''' | |
| − | + | | style="width:20%"|'''Lines of code''' | |
| − | + | | style="width:20%"|'''Person years''' | |
| − | + | | style="width:20%"|'''Price tag at 100k per person year''' | |
| − | + | |- | |
| − | + | | Slicer | |
| − | + | | align="right" | 55 | |
| − | + | | align="right" |604,093 | |
| − | + | | align="right" |165 | |
| + | | align="right" |$16,533,269 | ||
| + | |- | ||
| + | | KWW | ||
| + | | align="right" | 2 | ||
| + | | align="right" |189,635 | ||
| + | | align="right" |49 | ||
| + | | align="right" |$ 4,925,809 | ||
| + | |- | ||
| + | | VTK | ||
| + | | align="right" | 85 | ||
| + | | align="right" |1,385,777 | ||
| + | | align="right" |398 | ||
| + | | align="right" |$39,750,538 | ||
| + | |- | ||
| + | | ITK | ||
| + | | align="right" | 80 | ||
| + | | align="right" |727,275 | ||
| + | | align="right" |202 | ||
| + | | align="right" |$20,173,333 | ||
| + | |- | ||
| + | | CMAKE | ||
| + | | align="right" |25 | ||
| + | | align="right" |217,077 | ||
| + | | align="right" |57 | ||
| + | | align="right" |$ 5,680,460 | ||
| + | |- | ||
| + | | TEEM | ||
| + | | align="right" | 3 | ||
| + | | align="right" |114,065 | ||
| + | | align="right" |29 | ||
| + | | align="right" |$ 2,890,946 | ||
| + | |- | ||
| + | |style="background:#b3b3e7; color:black" |Total | ||
| + | | style="background:#b3b3e7; color:black" align="right" |250 | ||
| + | | style="background:#b3b3e7; color:black" align="right" |3,237,922 | ||
| + | | style="background:#b3b3e7; color:black" align="right" |900 | ||
| + | | style="background:#b3b3e7; color:black" align="right" |$ 89,954,355 | ||
| + | |} | ||
Latest revision as of 17:05, 21 November 2019
Home < Announcements:Slicer3.2Introduction
Links to Latest Slicer 3.2 Binaries
64-bit Linux: http://www.na-mic.org/Slicer/Download/Release/linux-x86_64/Slicer3-3.2.2008-08-08-linux-x86_64.tar.gz
32-bit Linux: http://www.na-mic.org/Slicer/Download/Release/linux-x86/Slicer3-3.2.2008-08-08-linux-x86.tar.gz
Darwin PPC: http://www.na-mic.org/Slicer/Download/Release/darwin-ppc/Slicer3-3.2.2008-08-08-darwin-ppc.tar.gz
Darwin x86: http://www.na-mic.org/Slicer/Download/Release/darwin-x86/Slicer3-3.2.2008-08-08-darwin-x86.tar.gz
Windows: http://www.na-mic.org/Slicer/Download/Release/win32/Slicer3-3.2.2008-08-08-win32.exe
The community of Slicer developers is proud to announce the release of Slicer 3.2. This effort is the culmination of hundreds of person years and tens of millions of dollars of effort [1]. Slicer leverages state of the art open-source toolkits for visualization, medical image analysis, software process, and other tools for processing and accessing data (for more information see the description of the NA-MIC Kit). Slicer offers these capabilities as part of the open-source framework known as the NA-MIC Kit, which facilitates on-going research in biomedical computing, supports commercialization through NA-MIC Kit components, and provides a spectrum of capabilities suitable for researchers with varying levels of computer skills.
|
Slicer 3.2 is a general purpose biomedical computing application with extensive built-in visualization and analysis capabilities, accessible through an easy to use graphical interface. For advanced users, Slicer may be extended at run-time with user-defined plug-in modules. Release candidates for this application will be available the first week of June 2008. This new release contains hundreds of changes to the software. Highlights include:
Click here to download different versions of Slicer3 and find pointers to the source code, mailing lists and bug tracker. Click here for a FAQ. Please note that Slicer continues to be a research package and is not intended for clinical use. Testing of functionality is an ongoing activity with high priority, however, some features of Slicer3 are not fully tested. |
Integrated Volume Rendering: View of the abdominal atlas Bone and large vessels are volume rendered. |
Highlights
- Slicer v3.2 - New and Improved Feature Highlights
Volume Rendering: Fully integrated volume rendering with cropping for easy exploration of volumetric data
Implicit Slice Widget: An interactive tool for specifying oblique views (part of the VTK widget family).
The Lightbox: Functionality for displaying cross-sectional data in columns and rows.
Scenesnapshots allow to capture and recover all parameters of a scene.
Interactive Editor: This new module allows interactive segmentation with robust 2D and 3D algorithms
IO: Improved IO capabilities include support for DICOM, NRRD, NIFTI, Tiff, JPG, Freesurfer, FITS and a number of other formats
Glyphs on corticospinal tract:
New Diffusion Imaging infrastructure includes, dicom import, gradient editor, visualiztionRemote Data Handling allows uploads and downloads from image informatics frameworks such as BIRN, and XNAT
IGT Toolkit to enable research in Image Guided Therapies
EM Segmenter: A configurable image segmentation tool that uses intensity distributions along with atlas information
Quality Software Process The NA-MIC Kit employs a test-driven software development process. Slicer3 also has a new build process.
Loadable Modules allows developers to provide modules to users outside of the build process, resulting in a smaller installed footprint and greater flexibility to end-users.
Slicer in Numbers
The numbers in this table represent the components of the NA-MIC kit. Slicer 3 is based on the NA-MIC kit.
Source: http://www.ohloh.org
Captured on July 31 2008. The numbers in the column entitled "lines of code" are hard numbers. The other two columns are estimates based on some assumptions. Please see the Ohloh website for an explanation of how the numbers were computed.
| Package | Approx. # Developers | Lines of code | Person years | Price tag at 100k per person year |
| Slicer | 55 | 604,093 | 165 | $16,533,269 |
| KWW | 2 | 189,635 | 49 | $ 4,925,809 |
| VTK | 85 | 1,385,777 | 398 | $39,750,538 |
| ITK | 80 | 727,275 | 202 | $20,173,333 |
| CMAKE | 25 | 217,077 | 57 | $ 5,680,460 |
| TEEM | 3 | 114,065 | 29 | $ 2,890,946 |
| Total | 250 | 3,237,922 | 900 | $ 89,954,355 |