Documentation/Nightly/Modules/PredictLesions
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: LesionSegmentation | |||
|
Module Description
This module is used to segment white matter lesions using an existing segmentation model. In order to use this tool your data must include a T1, T2, FLAIR, and brain mask. All data must be preprocessed including intra-subject co-registration, AC-PC alignment, bias correction, consistent spacing between sequences, and brain mask creation.
Use Cases
- Segmenting Lesions based on an existing model.
In order to perform segmentations you must first have access to an existing model (a model trained on 1.5T lupus data is available [|here]) and the images that were used for intensity standardization when training that model (the images for the lupus model are available [|here]. Performing a segmentation can then be done by loading the T1, T2, FLAIR, and brain mask for the subject you would like to segment, the reference T1, T2, FLAIR, and brain mask for intensity standardization, and the segmentation model. (Image of loading data to go here)
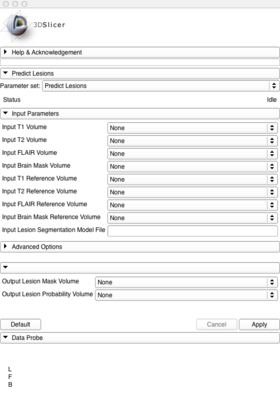
Navigate to Modules->Segmentation->LesionSegmentation->PredictLesions, then populate the appropriate parts of the PredictLesions input panel: (Image of populated input panel goes here.)
Then create a new volume (and a name, if you like) for the ouput lesion mask and the output probability volume. (Image of input panel with new outputs goes here)
Then press "Run" and wait approximately twenty minutes. The Lesion Probability Volume output will be an image where each voxel contains a value from 0 to 100 representing the percent chance that voxel is a lesion. The lesion mask contains zeros and ones and is a version of the Lesion Probability Volume thresholded at the inputLesionThreshold value which defaults to 70. (Image of Lesion Probability Volume) (Image of Lesion Mask)
If you navigate to the Volumes tab and select the Lesion Probability Volume you can then adjust the lower threshold to a value between 1-100. This effectively trades-off between sensitivity and specificity. (Image of adjusting the threshold)
Tutorials
Coming soon!
Panels and their use
|
Similar Modules
References
- Scully M, Anderson B, Lane T, Gasparovic C, Magnotta V, Sibbitt W, Roldan C, Kikinis R and Bockholt HJ (2010) An automated method for segmenting white matter lesions through multi-level morphometric feature classification with application to lupus. Front. Hum. Neurosci. doi:10.3389/fnhum.2010.00027
http://frontiersin.org/neuroscience/humanneuroscience/paper/10.3389/fnhum.2010.00027/
Information for Developers
| Section under construction. |