Documentation/4.6/Modules/Resampling
|
For the latest Slicer documentation, visit the read-the-docs. |
This page discusses Resampling of image and model data. See here for an overview of registration methods.
Contents
Transforms and how they are displayed in 3DSlicer
Slicer displays images in physical space, using information on Image Spacing (voxel size), image origin, coordinate system and the direction of the coordinate axes. This information is usually contained in the image header. Some of it can be seen in the Volumes module under Volume Information. If the same image appears differently in different software, the reason is most likely different use/interpretation of this header information.
A transform describes a relocation from one physical orientation into another physical orientation. All registration routines produce such a transform, but it may be optional to the user to just keep the reoriented image and not the transform. Note that if you keep the transform, you can access it via the Data module, you can save it to a text (ITK) file and apply it to other images. There are several modules available just for the task applying such transforms to an image, a process usually referred to as resampling.
Note there was a major upgrade to the support of nonlinear transforms added to nightly builds after v.4.3.1: non-rigid (BSpline) transforms are now supported for immediate visualization in the same fashion as linear (rigid to affine) transforms, i.e. placing a data object (image volume, label map, model, fiducial) within a transform will show in the slice and 3D views the updated position (drag image node onto the transform node in the Data module hierarchy). So for the newer version both transform types are handled/visualized the same, while version 4.3. and earlier distinguishes between linear (rigid to affine) and nonlinear (BSpline) transforms and does not show non-rigid transforms interactively but requires a resampling:
For version 4.3 and earlier: Slicer Registration Modules offer 2 types of output: an output transform and/or an output volume:
| Method | Output 1 | Output 2 |
| Rigid to Affine Registration | Output Transform | Output Volume |
| B-Spline Registration or combined Affine+B-Spline Registration | Output Transform | Output Volume |
- This result can be seen/viewed immediately in Slicer
- This result requires resampling first, e.g. using Resample Scalar/Vector/DWI Volume module or Resample Image (BRAINS) module
So the effect of a linear transform is therefore visible right away. Nonlinear (BSpline) transforms are not visualized on the fly and must be resampled to see. Hence if you calculate a BSpline transform inside BRAINSfit and then place a volume inside this transform, you will not see an effect, because the Viewer can apply only linear transforms directly. So to see a nonrigid transform you must resample the image. We therefore recommend to always select an output volume when running a nonrigid registration. To apply a transform and resample an image see the resampling via a spatial transformsection below.
Nesting & Hardening Transforms
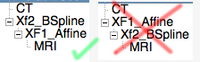
"Nesting" transforms is a term we use to describe the joining / concatenation of multiple transforms. This is supported in Slicer to some degree. The important thing to remember is that the order matters, i.e. A*B is not the same as B*A. For example, a typical application is that you first perform a rigid or affine registration, followed by a separate nonrigid BSpline registration. Although you can combine both in one, there are cases where it is useful to separate the two, e.g. if robustness is an issue and there is a chance the linear portion might fail, or for visualization, when you want to look at the deformations incurring from the BSpline, you need to have them separate from the global "bulk" transform to see detail. So in this case you need to nest the Affine inside the BSpline, not vice versa. If this seems counter-intuitive, remember that hierarchies are read/applied from the inside out, i.e. the first transform applied is the Affine and then the BSpline. So the arrangement within the data-module should look as shown on the image on the right (example of an MRI registered to a CT in 2 steps of first affine, then BSpline).
Hardening is a term used to describe applying a transform or effect to a dataset. In the Data module a right-click onto a node produces a context-menu, which lists Harden Transform as an option if a nesting is present. Two important points to remember about this option:
- hardening is not the same as resampling: in case of linear transforms, the new orientation is added to the physical orientation stored in the image header, without actually resampling the data itself. This is preferable in most cases as it avoids data degradation from interpolation artifacts associated with resampling. If you do need the transformed dataset resampled, use one of the resampling modules described below.
- hardening is currently supported for volumes inside BSpline transforms, but not for the transform itself, i.e. the image on the right shows which hardening will work and which will not. Hardening cannot currently be used to concatenate linear and BSpline or multiple BSpline transforms.
What we mean by Resampling
Resampling builds a new dataset (image, surface, fiducials etc.) from an existing one, but with a different orientation, resolution, field of view or aspect ratio. For example the last step in registering two images consists of two main steps: finding the transform and resampling according to this transform. So the last step in registration will be to resample the moving data according to a spatial transform function, and thereby generate a new and aligned image. Or changing the voxel size to something larger or smaller involves resampling.
See Wikipedia for a more detailed definition.
Interpolation is the process of estimating the value of the data based on surrounding values. This is necessary because spatial realignment is unlikely to be in exact multiples of voxel sizes. Please pay attention to selecting the proper interpolation method for your data-type. For more detail on interpolation we recommend this Wikipedia article.
Resampling in Place: Change Resolution or Field of View
- The Crop Volume module ("Modules:All Modules" menu) lets you clip away the image data outside an interactively defined 3D box. Such cropping is very useful to focus automated processing (e.g. registration, segmentation) onto a region of interest or reduce memory usage and speed up performance. The cropping includes a resampling that allows to change the resolution (spacing) of the result image at the same time.
- The Resample Scalar Volume module ("Modules:All Modules" menu) changes resolution (spacing) of an image, allowing several interpolation options for different data types. This is the method of choice if you wish to increase or decrease the number of voxels per mm or make the voxel size isotropic. You need to specify the desired voxel size in mm. If you do not know the current voxel size of your image, go to the Volume Information tab in the Volumes module.
- The Cast Scalar Volume module ("Modules:All Modules" menu) lets you change the underlying datatype of your image, e.g. change from floating point to an integer or vice versa. This will not perform an interpolation, since the number of image voxels stays the same, but may be necessary to prepare the image for some modules or to reduce the image (file) size. For example an image stored as float will need twice as much disk space than one stored as short and four times as much as a char.
- The Model To Label Map module ("Modules:Surface Models" menu) will do the inverse of the Model Maker and generate a binary 3D labelmap image with non-zero values at the location of the surface. This will let you bring the information from surface models back into an implicit image format. You set the resampling options, i.e. the resolution of the result via a reference image. Note that it will not fill a closed model, but produce points at the surface only.
- The Orient Scalar Images module ("Modules:Converters" menu) will sample the image data along a specified orientation scheme, such as axial/sagittal/coronal planes or RAS or LPI etc. Note that the slices are not interpolated, they are just reordered and/or permuted. The resulting volume will cover the original volume. Because the display is in physical space, you will not see an effect of this resampling in the viewer.
- The Volumes module ("Modules:Volumes" menu) lets you adjust the physical voxel size of an image (Info Tab), but will not generate a new volume. The changes will take effect immediately and you should see adjustments in your slice views. To obtain a new resampled volume, use the Resample Scalar Volume module described above.
- The Resample Image (BRAINS) module ("Modules:Registration" menu) lets you resample your image to a new resolution (specified implicitly via a reference image), with choice of interpolation method and output datatype. To resample in place leave the Warp by Transform menu on None.
Resampling via a spatial transform
- The Resample Image (BRAINS) module ("Modules:Registration" menu) supports applying linear transforms and deformation fields to an image, with choice of interpolation method and output datatype.
- The Resample Scalar/Vector/DWI Volume module ("Modules:All Modules" menu) sends both scalar and vector images through a transform. Several interpolation options.
- The Harden Transforms function (context menu via the right mouse button) in the Data module can also be used to resample an image or fiducial set through a linear transform.
- The AC-PC Transform module ("Modules:Registration:Specialized" menu) lets you realign your brain image along the anterior-posterior commissure and interhemispheral midline. You (manually) select fiducials to define these points. An output transform is generated that you can then apply to the image using the resampling tools described here.
- The main registration modules all contain a resampling option, i.e. they offer to produce a direct result volume, which includes a resampling step. In most cases the interpolation is linear. If you wish for more control over how the resampling is performed (e.g. select a different interpolator or output size or voxel size), select the registration module's option to generate an output/saved transform and then use one of the dedicated resampling modules described here to generate the new volume. The exception are modules which do not (yet) offer a transform output (e.g. Fast Nonrigid BSpline); for those you must use the module-specific resampling options to generate a result image.
- The Expert Automated Registration module ("Modules:All Modules" menu) offers pure resampling from a given transform: select None for initialization and None for registration, specify the transform to apply under Load Transform and set the interpolation type under Advanced Registration Parameters.
Resampling Vector- and Tensor-Data
- The Resample ResampleScalar/Vector/DWI Volume module ("Modules:All Modules" menu) is the method of choice to realign scalar, vector or diffusion weighted data along a given transform. It supports both linear and nonlinear transforms as well as deformation fields. Note that tensor data, such as DTI has a separate volume (below).
- The Resample DTI Volume module ("Modules:All Modules" menu) is designed specifically for reorienting diffusion tensor MRI data. Note the difference to DWI above. Use this for DTI but not for DWI. It supports both linear and nonlinear transforms as well as deformation fields. Note that simply sending each component of a DTI tensor through the transform separately would yield an incorrect result. This module will transform the vector/tensor data correctly.