Documentation/4.4/Modules/T1 Mapping
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: T1_Mapping | |||||||
|
Module Description
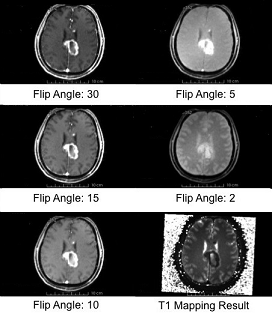
T1 mapping estimates effective tissue parameter maps (T1) from multi-spectral FLASH MRI scans with different flip angles.
Use Cases
- Take multi-spectral FLASH images with an arbitrary number of flip angles as input, and estimate the T1 values of the data for each voxel.
- Read repetition time(TR), echo time(TE) and flip angles from the Dicom header directly.
- Prostate, brain, head & neck, cervix, breast and etc.
Tutorials
Panels and their use
|
Similar Modules
References
Information for Developers
| Section under construction. |
Source code: https://github.com/stevedaxiao/T1_Mapping