Difference between revisions of "Modules:VMTKEasyLevelSetSegmentation"
(Created page with 'Return to Slicer 3.4 Documentation Gallery of New Features __NOTOC__ ===Module Name=== VMTKEasyLevelSetSegmentation…') |
|||
| (21 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | [[Documentation-3. | + | [[Documentation-3.6|Return to Slicer 3.6 Documentation]] |
| − | [[Announcements:Slicer3. | + | [[Announcements:Slicer3.6|Gallery of New Features]] |
__NOTOC__ | __NOTOC__ | ||
===Module Name=== | ===Module Name=== | ||
| − | VMTKEasyLevelSetSegmentation | + | '''VMTKEasyLevelSetSegmentation''', part of the [[Image:Slicervmtk logo.png||150px]] collection |
{| | {| | ||
| − | |[[Image: | + | |-valign="top" |
| − | |[[Image: | + | |[[Image:Easylevelsetgui.png|thumb|280x280px|Main GUI of VMTKEasyLevelSetSegmentation.]] |
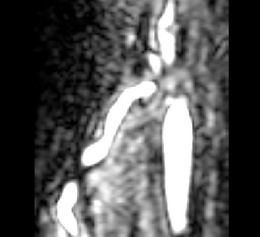
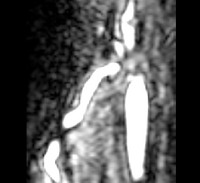
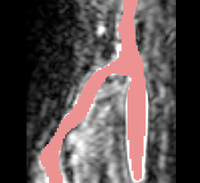
| − | |[[Image: | + | |[[Image:Avf_orig_axial.png|thumb|260x260px|Axial view of a contrast-enhanced three-dimensional image acquired from a MR scanner, showing an AV fistula in an arm. (Courtesy of L. Antiga, Bioengineering Department, Mario Negri Institute, Ranica (BG), Italy and N.R. Planken, Academisch Ziekenhuis Maastricht, the Netherlands.)]] |
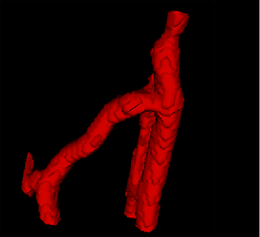
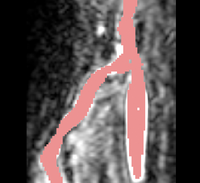
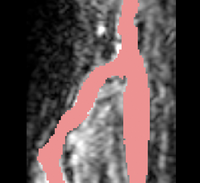
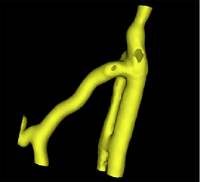
| − | |[[Image: | + | |[[Image:Avf_3d_initialization.png|thumb|260x260px|A surface model representation of the segmentation after the initialization step.]] |
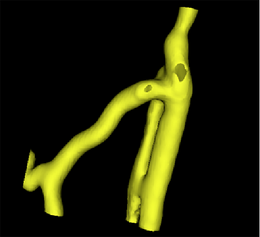
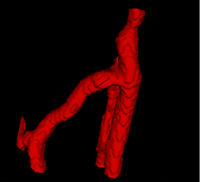
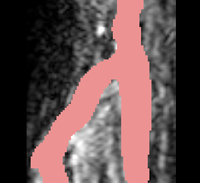
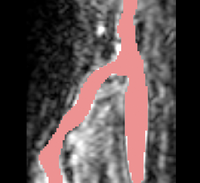
| + | |[[Image:Avf_3d_evolution_new.png|thumb|260x260px|A surface model showing the segmentation after the evolution was performed.]] | ||
|} | |} | ||
| Line 27: | Line 28: | ||
===Module Description=== | ===Module Description=== | ||
| − | This module provides level-set segmentation in 3D Slicer using selected methods of the '''level-set segmentation process of the Vascular Modeling Toolkit''' (http://www.vmtk.org). The module targets easy-to-use manual segmentation of tubular and blob-like structures. The methods provided by this module are ''Fast Marching Upwind Gradient Initialization'' and ''Geodesic Active Contours''. | + | This module provides level-set segmentation in 3D Slicer using selected methods of the '''level-set segmentation process of the Vascular Modeling Toolkit''' (http://www.vmtk.org). The module targets easy-to-use manual segmentation of tubular and blob-like structures. The methods provided by this module are ''Fast Marching Upwind Gradient Initialization'' and ''Geodesic Active Contours'' and ''CURVES Evolution''. |
Segmentation using level sets consists of an initialization and an evolution step. The initialization step involves the description of a starting model within a region of interest. In the evolution step this initial deformable model then gets inflated to match the contours of the targeted volume. | Segmentation using level sets consists of an initialization and an evolution step. The initialization step involves the description of a starting model within a region of interest. In the evolution step this initial deformable model then gets inflated to match the contours of the targeted volume. | ||
| Line 39: | Line 40: | ||
''This module depends on the [[Modules:VMTKSlicerModule|VmtkSlicerModule]]: see [[Modules:VMTKSlicerModule|this page]] for installation notes.'' | ''This module depends on the [[Modules:VMTKSlicerModule|VmtkSlicerModule]]: see [[Modules:VMTKSlicerModule|this page]] for installation notes.'' | ||
| − | The VMTKEasyLevelSetSegmentation module can be installed | + | The VMTKEasyLevelSetSegmentation module can be installed using the '''3D Slicer extension wizard'''. The extension is called ''VMTKEasyLevelSetSegmentation''. |
| − | + | When the module was successfully installed, it is available within 3D Slicer's module selector inside the category ''Vascular Modeling Toolkit''. | |
| − | |||
| − | + | ===Examples, Use Cases=== | |
| − | |||
| − | |||
| − | |||
| − | + | The following screencasts/tutorials show different Use Cases of the VMTKEasyLevelSetSegmentation module. | |
| + | * '''Use Case #1''': Segmentation of a Cerebral Aneurysm using two Fiducial Points | ||
| + | {| | ||
| + | |-valign="top" | ||
| + | |[[Image:VMTKEasyLevelSetSegmentationAneurysmScreencast.png|thumb|280px|Watch the [[Modules:VMTKEasyLevelSetSegmentation_AneurysmTutorial|video]]..]] | ||
| + | |The segmentation of a cerebral aneurysm and its connecting vessels using the VMTKEasyLevelSetSegmentation module is shown in this screencast. Two fiducial points are used to border the segmented area inside the region of interest as input for the Fast Marching segmentation. | ||
| − | + | Label maps showing the evolved segmentation in comparison to the initial one are generated after initialization and evolution stage. | |
| + | |||
| + | Watch the video [[Modules:VMTKEasyLevelSetSegmentation_AneurysmTutorial|here]]. | ||
| + | |[[Image:VMTKEasyLevelSetSegmentationAneurysma_sliceviewers_initialized.png|thumb|450px|Slice viewers showing the initialization as a label map]] | ||
| + | [[Image:VMTKEasyLevelSetSegmentationAneurysma_sliceviewers_evolved.png|thumb|450px|Slice viewers showing a label map of the smoother surface after it was evolved]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | * '''Use Case #2''': Segmentation of the Coronary Arteries inside a Cardiac Blood-pool MRI using one Fiducial Point | ||
| + | {| | ||
| + | |-valign="top" | ||
| + | |[[Image:VMTKEasyLevelSetSegmentationCoronariesScreencast.png|thumb|280px|Watch the [[Modules:VMTKEasyLevelSetSegmentation_CoronariesTutorial|video]]..]] | ||
| + | |The segmentation of the coronary arteries inside an already vessel enhanced cardiac blood-pool MRI using the VMTKEasyLevelSetSegmentation module is shown in this tutorial. One fiducial is used to start the segmentation. The vessel enhancement was performed using the [[Modules:VMTKVesselEnhancement|VMTKVesselEnhancement]] module and a region of interest was selected. | ||
| + | |||
| + | Watch the video [[Modules:VMTKEasyLevelSetSegmentation_CoronariesTutorial|here]]. | ||
| + | |} | ||
| + | |||
| + | * '''Use Case #3''': Segmentation of Bronchi in a CT of the left Lung using one Fiducial Point | ||
| + | {| | ||
| + | |-valign="top" | ||
| + | |[[Image:VMTKEasyLevelSetSegmentationBronchiScreencast.png|thumb|280px|Watch the [[Modules:VMTKEasyLevelSetSegmentation_BronchiTutorial|video]]..]] | ||
| + | |This tutorial shows the segmentation of bronchi in a region of interest of the left lung in a CT volume using the VMTKEasyLevelSetSegmentation module. One fiducial is used to start the Fast Marching segmentation. | ||
| − | + | The over-segmentation shown by leakage to the surrounding tissue, is a disadvantage of the current method and has to be fixed.. | |
| − | + | Watch the video [[Modules:VMTKEasyLevelSetSegmentation_BronchiTutorial|here]]. | |
| + | |} | ||
| − | * '''Use Case # | + | * '''Use Case #4''': Evolving an already existing initial segmentation using the evolution stage only |
{| | {| | ||
|-valign="top" | |-valign="top" | ||
| − | |[[Image: | + | |[[Image:VMTKEasyLevelSetSegmentationAneurysmOnlyEvolutionScreencast.png|thumb|280px|Watch the [[Modules:VMTKEasyLevelSetSegmentation_AneurysmEvolutionOnlyTutorial|video]]..]] |
| − | | | + | |This tutorial shows how to use the VMTKEasyLevelSetSegmentation module to evolve an initial segmentation of a cerebral aneurysm which was obtained using a different module (f.e. using the original [[Modules:VMTKLevelSetSegmentation|VMTKLevelSetSegmentation]]) to the gradient ridges of the original volume. The initial segmentation is a binary image. |
| + | |||
| + | Watch the video [[Modules:VMTKEasyLevelSetSegmentation_AneurysmEvolutionOnlyTutorial|here]]. | ||
|} | |} | ||
| − | * ''' | + | |
| + | * '''Example segmentation''': The following images show results of an example segmentation using this tool. An AV fistula is segmented in a contrast-enhanced three-dimensional image acquired from a MR scanner. | ||
| + | First, we perform the initialization on a vessel enhanced version of the original image. A single click inside the vessel of interest and a threshold (here 0.1-1.0) are sufficient to obtain the first segmentation. | ||
{| | {| | ||
|-valign="top" | |-valign="top" | ||
| − | |[[Image: | + | |[[Image:Avf_orig_axial.png|thumb|200x200px|Axial view of the original image showing the AV fistula. (Courtesy of L. Antiga, Bioengineering Department, Mario Negri Institute, Ranica (BG), Italy and N.R. Planken, Academisch Ziekenhuis Maastricht, the Netherlands.)]] |
| − | | | + | |[[Image:Avf_frangi_multiscale.png|thumb|200x200px|The same image after multiscale vessel enhancement using the [[Modules:VMTKVesselEnhancement|VMTKVesselEnhancement]] module.]] |
| + | |[[Image:Avf_fm_label_map.png|thumb|200x200px|Resulting Fast Marching segmentation after single seed initialization inside the vessel tree shown as a label map.]] | ||
| + | |[[Image:Avf_3d_initialization.png|thumb|200x200px|A surface model representation of the segmentation after the initialization step.]] | ||
|} | |} | ||
| − | + | This initialization can be optimized during the evolution stage. The segmentation gets deformed and attaches to the gradient ridges (a.k.a. the vessel borders). The deformation behaviour can be configured using different weights. | |
{| | {| | ||
|-valign="top" | |-valign="top" | ||
| − | |[[Image: | + | |[[Image:Avf_gac_20it_-100_0_-100.png|thumb|200x200px|Label map representation of the segmentation after the evolution stage with maximal inflation.]] |
| − | |The | + | |[[Image:Avf_gac_20it_0_100_0.png|thumb|200x200px|Label map representation of the segmentation after the evolution stage with maximal curvature.]] |
| + | |[[Image:Avf_gac_20it_0_0_-100.png|thumb|200x200px|Label map representation of the segmentation after the evolution stage with maximal attraction to the gradient ridges.]] | ||
| + | |[[Image:Avf_gac_20it_0_60_-100.png|thumb|200x200px|The best evolution result is usually obtained by choosing medium curvature (~60) and maximal attraction.]] | ||
| + | |[[Image:Avf_3d_evolution_new.png|thumb|200x200px|A surface model representation of the segmentation after the evolution step. The model appears much smoother than the output after the initialization step.]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | ===Tutorials=== | ||
| + | The following tutorials demonstrate the use of VMTKEasyLevelSetSegmentation in a pipeline of VMTK modules to extract the centerlines of coronary arteries. | ||
| + | |||
| + | {| | ||
| + | |[[Image:Vmtkaftercenterline.png|thumb|260x260px|Step-by-step online tutorial: Segmentation of the Right Coronary Artery using VMTK in 3D Slicer. View online [[Modules:VMTK_in_3D_Slicer_Tutorial:_Coronary_Artery_Centerline_Extraction|here]].]] | ||
| + | |[[Image:VMTKCenterlines_Tutorial_first_slide.png|thumb|280x280px|Step-by-step tutorial including example dataset: The first slide of the Centerline Extraction tutorial. Download [http://www.na-mic.org/Wiki/images/4/40/TutorialVMTKCoronariesCenterlinesMRI_Winter2010AHM.pdf here].]] | ||
|} | |} | ||
===Quick Tour of Features and Use=== | ===Quick Tour of Features and Use=== | ||
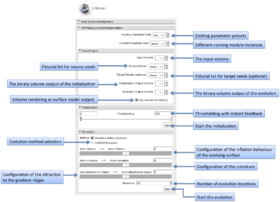
| − | The VMTKEasyLevelSetSegmentation | + | The graphical user interface of the VMTKEasyLevelSetSegmentation module consists of four panels. |
| + | [[Image:Easylevelsetgui.png|thumb|500x500px|The GUI of VMTKEasyLevelSetSegmentation. Click to enlarge.]] | ||
| + | |||
| + | * Main module panel: | ||
| + | Parameter presets exist for different use-cases. These get loaded from the MRML file ''presets.xml'' inside the module directory and can be edited. | ||
| + | The current parameters are stored to the MRML node of the module and therefore it is possible to work with different parameter sets. | ||
| + | |||
| + | * I/O panel: | ||
| + | The input volume and the output volumes for the initialization and evolution stage can be selected using this panel. | ||
| + | The output volume of the initialization stage is used as an input for the evolution stage. | ||
| + | Fiducial lists can be configured as source and target seeds. If the selected fiducial lists are equal, target seeds will be ignored. This is useful for the segmentation of vessel trees. | ||
| + | A switch to choose between 3D visualization using models generated by Marching Cubes or Volume Rendering is available. | ||
| + | |||
| + | * Initialization panel: | ||
| + | The threshold slider is used as a parameter for the Fast Marching Upwind Gradient Image Filter and updates the slice viewers during change for visualization. | ||
| + | * Evolution panel: | ||
| + | The module supports to choose between Geodesic Active Contours and CURVES evolution methods using a checkbox. | ||
| + | Different weights are available to configure the inflation of the initial level-set. Is is possible to adjust the importance of inflation, curvature shape and attraction to the gradient ridges. | ||
| + | The number of iterations defines how many steps of evolution are performed. | ||
The module supports the 3D Slicer built-in ''Undo'' function. | The module supports the 3D Slicer built-in ''Undo'' function. | ||
| − | All generated binary volumes and used fiducials are saved as MRML nodes in the current MRML scene. Therefore these can be exported or saved. | + | All generated models, binary volumes and used fiducials are saved as MRML nodes in the current MRML scene. Therefore these can be exported or saved. |
== Development == | == Development == | ||
| + | |||
| + | ===Notes from the Developer(s)=== | ||
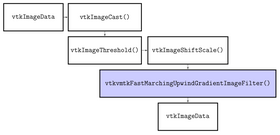
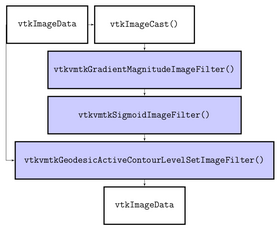
| + | The algorithms provided by VMTK and used by this module are C++ classes implemented using VTK and ITK. In particular the module uses the Fast Marching Upwind Gradient Image Filter (see http://math.berkeley.edu/~sethian/Explanations/fast_marching_explain.html) for initialization and the Geodesic Active Contours (see http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.21.2196&rep=rep1&type=pdf) and CURVES (see http://www.spl.harvard.edu/publications/item/view/179) level set filters for evolution. | ||
===Dependencies=== | ===Dependencies=== | ||
| − | This module depends on the VMTK libraries which are provided in the [[Modules:VMTKSlicerModule|VmtkSlicerModule]]. Therefore the [[Modules:VMTKSlicerModule|VmtkSlicerModule]] has to be installed before the | + | This module depends on the VMTK libraries which are provided in the [[Modules:VMTKSlicerModule|VmtkSlicerModule]]. Therefore the [[Modules:VMTKSlicerModule|VmtkSlicerModule]] has to be installed before the VMTKEasyLevelSetSegmentation module can be used. |
===Known bugs & Usability issues=== | ===Known bugs & Usability issues=== | ||
| Line 98: | Line 163: | ||
===Source code & documentation=== | ===Source code & documentation=== | ||
{| | {| | ||
| − | |[[Image: | + | |[[Image:Easylevelset_classdiagram.png|thumb|280x280px|Class diagram showing the separation of GUI and logic]] |
| − | |[[Image: | + | |[[Image:Fastmarching_pipeline.png|thumb|280x280px|Flow chart showing the VTK/VMTK pipeline of the Fast Marching algorithm]] |
| − | |[[Image: | + | |[[Image:Gac_pipeline.png|thumb|280x280px|Flow chart showing the VTK/VMTK pipeline of the Geodesic Active Contours algorithm]] |
|} | |} | ||
VMTKEasyLevelSetSegmentation is a Python Scripted Module. It follows the conventions of the Model View Controller pattern of slicer modules. This implies the separation of logic and GUI. | VMTKEasyLevelSetSegmentation is a Python Scripted Module. It follows the conventions of the Model View Controller pattern of slicer modules. This implies the separation of logic and GUI. | ||
| Line 106: | Line 171: | ||
The class ''VMTKEasyLevelSetSegmentationGUI'' derives from ''ScriptedModuleGUI'' and saves the current parameters to its own MRML node. The actual calls to the VMTK libraries as well as the Marching Cubes or Volume Rendering methods are performed in the class ''VMTKEasyLevelSetSegmentationLogic''. Several general supporting functions are outsourced in the class ''VMTKEasyLevelSetSegmentationHelper''. | The class ''VMTKEasyLevelSetSegmentationGUI'' derives from ''ScriptedModuleGUI'' and saves the current parameters to its own MRML node. The actual calls to the VMTK libraries as well as the Marching Cubes or Volume Rendering methods are performed in the class ''VMTKEasyLevelSetSegmentationLogic''. Several general supporting functions are outsourced in the class ''VMTKEasyLevelSetSegmentationHelper''. | ||
| − | The complete source code is available at a [http://www.nitrc.org/plugins/scmsvn/viewcvs.php/ | + | The complete source code is available at a [http://www.nitrc.org/plugins/scmsvn/viewcvs.php/VMTKEasyLevelSetSegmentation/?root=slicervmtklvlst NITRC SVN repository]. |
== More Information == | == More Information == | ||
| Line 114: | Line 179: | ||
===References=== | ===References=== | ||
| + | * Antiga L, Piccinelli M, Botti L, Ene-Iordache B, Remuzzi A and Steinman DA. An image-based modeling framework for patient-specific computational hemodynamics. Medical and Biological Engineering and Computing, 46: 1097-1112, Nov 2008. | ||
| + | * D. Hähn. Integration of the vascular modeling toolkit in 3d slicer. SPL, 04 2009. Available online at http://www.spl.harvard.edu/publications/item/view/1728. | ||
| + | * D. Hähn. Centerline Extraction of Coronary Arteries in 3D Slicer using VMTK based Tools. Master's Thesis. Department of Medical Informatics, University of Heidelberg, Germany. Feb 2010. | ||
* Piccinelli M, Veneziani A, Steinman DA, Remuzzi A, Antiga L (2009) A framework for geometric analysis of vascular structures: applications to cerebral aneurysms. IEEE Trans Med Imaging. In press. | * Piccinelli M, Veneziani A, Steinman DA, Remuzzi A, Antiga L (2009) A framework for geometric analysis of vascular structures: applications to cerebral aneurysms. IEEE Trans Med Imaging. In press. | ||
| − | |||
Latest revision as of 13:53, 20 April 2010
Home < Modules:VMTKEasyLevelSetSegmentationReturn to Slicer 3.6 Documentation
Module Name
VMTKEasyLevelSetSegmentation, part of the  collection
collection
General Information
Module Type & Category
Type: Scripted Module
Category: Segmentation, Extension
Authors, Collaborators & Contact
- Author: Daniel Haehn, University of Heidelberg
- Acknowledgments: Luca Antiga, Mario Negri Institute; Steve Pieper, Isomics Inc.
- Contact: Daniel Haehn, haehn@bwh.harvard.edu
Module Description
This module provides level-set segmentation in 3D Slicer using selected methods of the level-set segmentation process of the Vascular Modeling Toolkit (http://www.vmtk.org). The module targets easy-to-use manual segmentation of tubular and blob-like structures. The methods provided by this module are Fast Marching Upwind Gradient Initialization and Geodesic Active Contours and CURVES Evolution.
Segmentation using level sets consists of an initialization and an evolution step. The initialization step involves the description of a starting model within a region of interest. In the evolution step this initial deformable model then gets inflated to match the contours of the targeted volume.
This work is part of the NA-MIC VMTK Collaboration.
Official project page: http://www.vmtk.org/Main/VmtkIn3DSlicer
Usage
Installation
This module depends on the VmtkSlicerModule: see this page for installation notes.
The VMTKEasyLevelSetSegmentation module can be installed using the 3D Slicer extension wizard. The extension is called VMTKEasyLevelSetSegmentation.
When the module was successfully installed, it is available within 3D Slicer's module selector inside the category Vascular Modeling Toolkit.
Examples, Use Cases
The following screencasts/tutorials show different Use Cases of the VMTKEasyLevelSetSegmentation module.
- Use Case #1: Segmentation of a Cerebral Aneurysm using two Fiducial Points
 Watch the video.. |
The segmentation of a cerebral aneurysm and its connecting vessels using the VMTKEasyLevelSetSegmentation module is shown in this screencast. Two fiducial points are used to border the segmented area inside the region of interest as input for the Fast Marching segmentation.
Label maps showing the evolved segmentation in comparison to the initial one are generated after initialization and evolution stage. Watch the video here. |
- Use Case #2: Segmentation of the Coronary Arteries inside a Cardiac Blood-pool MRI using one Fiducial Point
 Watch the video.. |
The segmentation of the coronary arteries inside an already vessel enhanced cardiac blood-pool MRI using the VMTKEasyLevelSetSegmentation module is shown in this tutorial. One fiducial is used to start the segmentation. The vessel enhancement was performed using the VMTKVesselEnhancement module and a region of interest was selected.
Watch the video here. |
- Use Case #3: Segmentation of Bronchi in a CT of the left Lung using one Fiducial Point
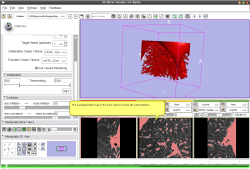 Watch the video.. |
This tutorial shows the segmentation of bronchi in a region of interest of the left lung in a CT volume using the VMTKEasyLevelSetSegmentation module. One fiducial is used to start the Fast Marching segmentation.
The over-segmentation shown by leakage to the surrounding tissue, is a disadvantage of the current method and has to be fixed.. Watch the video here. |
- Use Case #4: Evolving an already existing initial segmentation using the evolution stage only
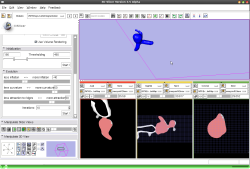 Watch the video.. |
This tutorial shows how to use the VMTKEasyLevelSetSegmentation module to evolve an initial segmentation of a cerebral aneurysm which was obtained using a different module (f.e. using the original VMTKLevelSetSegmentation) to the gradient ridges of the original volume. The initial segmentation is a binary image.
Watch the video here. |
- Example segmentation: The following images show results of an example segmentation using this tool. An AV fistula is segmented in a contrast-enhanced three-dimensional image acquired from a MR scanner.
First, we perform the initialization on a vessel enhanced version of the original image. A single click inside the vessel of interest and a threshold (here 0.1-1.0) are sufficient to obtain the first segmentation.
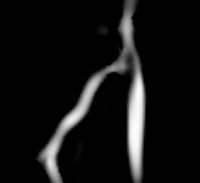 The same image after multiscale vessel enhancement using the VMTKVesselEnhancement module. |
This initialization can be optimized during the evolution stage. The segmentation gets deformed and attaches to the gradient ridges (a.k.a. the vessel borders). The deformation behaviour can be configured using different weights.
Tutorials
The following tutorials demonstrate the use of VMTKEasyLevelSetSegmentation in a pipeline of VMTK modules to extract the centerlines of coronary arteries.
 Step-by-step online tutorial: Segmentation of the Right Coronary Artery using VMTK in 3D Slicer. View online here. |
 Step-by-step tutorial including example dataset: The first slide of the Centerline Extraction tutorial. Download here. |
Quick Tour of Features and Use
The graphical user interface of the VMTKEasyLevelSetSegmentation module consists of four panels.
- Main module panel:
Parameter presets exist for different use-cases. These get loaded from the MRML file presets.xml inside the module directory and can be edited. The current parameters are stored to the MRML node of the module and therefore it is possible to work with different parameter sets.
- I/O panel:
The input volume and the output volumes for the initialization and evolution stage can be selected using this panel. The output volume of the initialization stage is used as an input for the evolution stage. Fiducial lists can be configured as source and target seeds. If the selected fiducial lists are equal, target seeds will be ignored. This is useful for the segmentation of vessel trees. A switch to choose between 3D visualization using models generated by Marching Cubes or Volume Rendering is available.
- Initialization panel:
The threshold slider is used as a parameter for the Fast Marching Upwind Gradient Image Filter and updates the slice viewers during change for visualization.
- Evolution panel:
The module supports to choose between Geodesic Active Contours and CURVES evolution methods using a checkbox.
Different weights are available to configure the inflation of the initial level-set. Is is possible to adjust the importance of inflation, curvature shape and attraction to the gradient ridges. The number of iterations defines how many steps of evolution are performed.
The module supports the 3D Slicer built-in Undo function.
All generated models, binary volumes and used fiducials are saved as MRML nodes in the current MRML scene. Therefore these can be exported or saved.
Development
Notes from the Developer(s)
The algorithms provided by VMTK and used by this module are C++ classes implemented using VTK and ITK. In particular the module uses the Fast Marching Upwind Gradient Image Filter (see http://math.berkeley.edu/~sethian/Explanations/fast_marching_explain.html) for initialization and the Geodesic Active Contours (see http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.21.2196&rep=rep1&type=pdf) and CURVES (see http://www.spl.harvard.edu/publications/item/view/179) level set filters for evolution.
Dependencies
This module depends on the VMTK libraries which are provided in the VmtkSlicerModule. Therefore the VmtkSlicerModule has to be installed before the VMTKEasyLevelSetSegmentation module can be used.
Known bugs & Usability issues
Follow this link to the VMTK in 3D Slicer bug tracker.
Source code & documentation
VMTKEasyLevelSetSegmentation is a Python Scripted Module. It follows the conventions of the Model View Controller pattern of slicer modules. This implies the separation of logic and GUI.
The class VMTKEasyLevelSetSegmentationGUI derives from ScriptedModuleGUI and saves the current parameters to its own MRML node. The actual calls to the VMTK libraries as well as the Marching Cubes or Volume Rendering methods are performed in the class VMTKEasyLevelSetSegmentationLogic. Several general supporting functions are outsourced in the class VMTKEasyLevelSetSegmentationHelper.
The complete source code is available at a NITRC SVN repository.
More Information
Acknowledgment
This work was funded by a grant of the Thomas-Gessmann Foundation part of the Founder Federation for German Science.
References
- Antiga L, Piccinelli M, Botti L, Ene-Iordache B, Remuzzi A and Steinman DA. An image-based modeling framework for patient-specific computational hemodynamics. Medical and Biological Engineering and Computing, 46: 1097-1112, Nov 2008.
- D. Hähn. Integration of the vascular modeling toolkit in 3d slicer. SPL, 04 2009. Available online at http://www.spl.harvard.edu/publications/item/view/1728.
- D. Hähn. Centerline Extraction of Coronary Arteries in 3D Slicer using VMTK based Tools. Master's Thesis. Department of Medical Informatics, University of Heidelberg, Germany. Feb 2010.
- Piccinelli M, Veneziani A, Steinman DA, Remuzzi A, Antiga L (2009) A framework for geometric analysis of vascular structures: applications to cerebral aneurysms. IEEE Trans Med Imaging. In press.