Home < Documentation < 4.1 < Modules < MultiVolumeImporter
Introduction and Acknowledgements
|
This work is supported by NA-MIC, NAC, NCIGT, and the Slicer Community. This work is partially supported by the following grants: P41EB015898, P41RR019703, R01CA111288 and U01CA151261.
Author: Andrey Fedorov, Jean-Cristophe Fillion Robin, Julien Finet, Steve Pieper, Ron Kikinis
Contact: Andrey Fedorov <email>fedorov@bwh.harvard.edu</email>
|
| National Center for Image Guided Therapy (NCIGT)
|
| National Alliance for Medical Image Computing (NA-MIC)
|
| Surgical Planning Laboratory (SPL)
|
| Neuroimage Analysis Center (NAC)
|
|
|
Module Description
This module provides support for importing multivolume (multiframe) data.
Use Cases
Most frequently used for these scenarios:
- importing a DICOM dataset that contains multiple frames that can be separated based on some tag (e.g., DCE MRI data, where individual temporally resolved frames are identified by Trigger Time tag (0018,1060)
- importing multiple frames defined in the same coordinate frame, saved as individual volumes in NRRD, NIfTI, or any other image format supported by 3D Slicer
Tutorials
- Sample datasets are available:
Panels and their use
- Basic settings
- Input directory: location of the input data (either in DICOM format, or as a collection of frames)
- Output node: MultiVolume node that will keep the loaded data. You need to create a new node or select and existing one when importing the data.
- Input data type: multivolume data can be imported from an appropriate DICOM series, or from a collection of individual frames saved in one of the Slicer-supported volume file formats. Supported options are:
- DICOM 4D DCE MRI: individual frames will be separated from the DICOM series by (0018,1060) tag (Trigger Time)
- DICOM variable TE MRI: (0018,0081) (Echo Time)
- DICOM variable FA MRI: (0018,1314) (Flip Angle)
- DICOM variable TR MRI: (0018,0080) (Repetition Time)
- User-defined DICOM: the tag specified by the user in the Advanced settings panel will be used to separate individual frames.
- User-defined non-DICOM: the directory is expected to contain a list of individual frames saved in a image volume format supported by 3D Slicer, such as NRRD or NIfTI.
- IMPORTANT If you use User-defined non-DICOM input data type, the frames will be sorted based on the alphanumerical order of the frame filenames. If you have more than 10 frames, you should name them as follows to make sure they are ordered correctly. Correct naming: frame001.nrrd, frame002.nrrd, ..., frame023.nrrd, ..., frame912.nrrd. Incorrect naming: frame1.nrrd, frame2.nrrd, ..., frame14.nrrd, ..., frame1045.nrrrd.
|
|
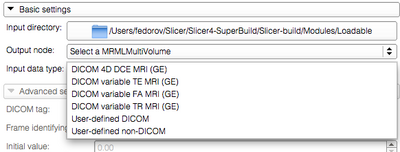 Basic settings panel with the "Input data type" dropbox expanded |
- Advanced settings: contains elements that can be changed by the user when Input data type selected is either user-defined DICOM or user-defined non-DICOM
- DICOM tag: in all modes, shows the DICOM tag that will be used to separate individual frames/volumes in the DICOM series. This field does not have meaning when the input data type is non-DICOM!
- Frame identifying units: automatically populated for pre-defined tags. Needs to be defined for other input data types.
- Initial value: values of the frame-identifying units for non-DICOM data type.
|
|
|
- Import button: once the panels are populated with the appropriate settings, hit this button to import the dataset into Slicer. Note that depending on the size of the data this operation can take significant time, so be patient!
Similar Modules
Once the multivolume dataset is loaded, it can be viewed and further explored using MultiVolumeExplorer module.
References
Information for Developers
Known issues that won't be resolved
- bug 1841: multivolumes that contain less than 7 frames will not be saved correctly and the scene containing such objects will not be loaded correctly by Slicer (limitation of the NRRD IO for DWI)
Features to be implemented
- integrate MV importer as a DICOM module plugin
Features under consideration
- should DWI node be a child under the MV hierarchy?
- import multivolumes from ITK 4d images