Difference between revisions of "Announcements:Slicer3.6"
Sylvainjaume (talk | contribs) (→Highlights: use a close-up screenshot) |
|||
| Line 28: | Line 28: | ||
Image:SlicerFiducialsGlyphs-3.4.jpg| [[Modules:Fiducials-Documentation-3.6|Fiducials]] (Nicole Aucoin) | Image:SlicerFiducialsGlyphs-3.4.jpg| [[Modules:Fiducials-Documentation-3.6|Fiducials]] (Nicole Aucoin) | ||
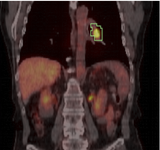
Image:PETCTFusion.png| [[Modules:PETCTFusion-Documentation-3.6 |PETCTFusion]] (Wendy Plesniak) | Image:PETCTFusion.png| [[Modules:PETCTFusion-Documentation-3.6 |PETCTFusion]] (Wendy Plesniak) | ||
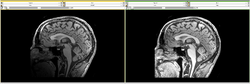
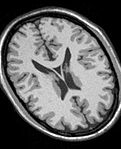
| − | Image: | + | Image:MRI_Bias_Field_Correction_Slicer3_close_up.png|[[Modules:MRIBiasFieldCorrection-Documentation-3.6|MRIBiasFieldCorrection]] correction of MRI intensity inhomogeneity i.e. bias field (Sylvain Jaume) |
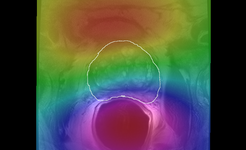
Image:N4 prostate bias ax.png| [[Modules:N4ITKBiasFieldCorrection-Documentation-3.6|N4 Bias Field Correction]] (Andriy Fedorov) | Image:N4 prostate bias ax.png| [[Modules:N4ITKBiasFieldCorrection-Documentation-3.6|N4 Bias Field Correction]] (Andriy Fedorov) | ||
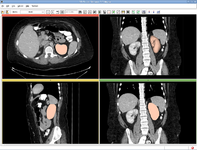
Image:RSSkidneyL.png| [[Modules:RobustStatisticsSeg-Documentation-3.6|Robust Statistical Segmentation]] (Yi Gao) | Image:RSSkidneyL.png| [[Modules:RobustStatisticsSeg-Documentation-3.6|Robust Statistical Segmentation]] (Yi Gao) | ||
Revision as of 19:01, 2 May 2010
Home < Announcements:Slicer3.6
Back to Documentation 3.6
Introduction
The community of Slicer developers is proud to announce the release of Slicer 3.6.
- Click here to download different versions of Slicer3 and find pointers to the source code, mailing lists and bug tracker. *Please note that Slicer continues to be a research package and is not intended for clinical use. Testing of functionality is an ongoing activity with high priority, however, some features of Slicer3 are not fully tested.
- The Slicer Training page provides a series of courses for learning how to use Slicer3. The portfolio contains self-guided presentation and sample data sets
The main slicer.org pages provide a guided tour to the application, training materials, and the development community. New users should start there because we try to keep the pages organized and up to date.
Highlights
- Slicer v3.6 - New and Improved Feature Highlights
The Interactive Editor can be used to create and edit label maps for quantitative analysis and surface model generation
Virtual Endoscopy (Steve Pieper)
Colors (Nicole Aucoin)
4D Image Viewer (Junichi Tokuda)
Fast Marching segmentation (Andriy Fedorov)
Mesh Contour Segmentation (Peter Karasev)
Crop Volume (Andriy Fedorov)
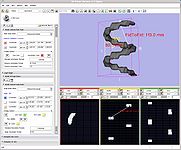
Measurements (rulers and angles) (Nicole Aucoin)
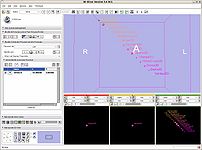
Affine registration (Casey Goodlett)
Fiducials (Nicole Aucoin)
PETCTFusion (Wendy Plesniak)
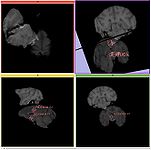
MRIBiasFieldCorrection correction of MRI intensity inhomogeneity i.e. bias field (Sylvain Jaume)
N4 Bias Field Correction (Andriy Fedorov)
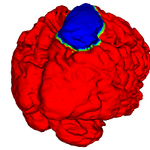
Robust Statistical Segmentation (Yi Gao)
Model Transform (Alex Yarmarkovich)
Resample Scalar/Vector/DWI Volume (Francois Budin)