Modules:ResampleVolumeBatch-Documentation-3.6
Return to Slicer 3.6 Documentation
Module Name
Resample Scalar Volume BatchMake
General Information
Module Type & Category
Type: CLI
Category: Filtering
Authors, Collaborators & Contact
- Author: Stephen Aylward and Julien Finet at Kitware Inc.
- Contact: stephen.aylward at kitware.com
Module Description
This module is the BatchMake version of the Resample Scalar Volume module. It can resample automatically every file in a the input directory with a specified scaling.
Resampling an image is an important task in image analysis. It is especially important in the frame of image registration. This module implements image resampling through the use of itk Transforms. This module uses an Identity Transform. The resampling is controlled by the Output Spacing. 'Resampling' is performed in space coordinates, not pixel/grid coordinates. It is quite important to ensure that image spacing is properly set on the images involved. The interpolator is required since the mapping from one space to the other will often require evaluation of the intensity of the image at non-grid positions. Several interpolators are available: linear, nearest neighbor and five flavors of sinc. The sinc interpolators, although more precise, are much slower than the linear and nearest neighbor interpolator. To resample label volumnes, nearest neighbor interpolation should be used exclusively.
See the Resample Volume documentation for more information.
Usage
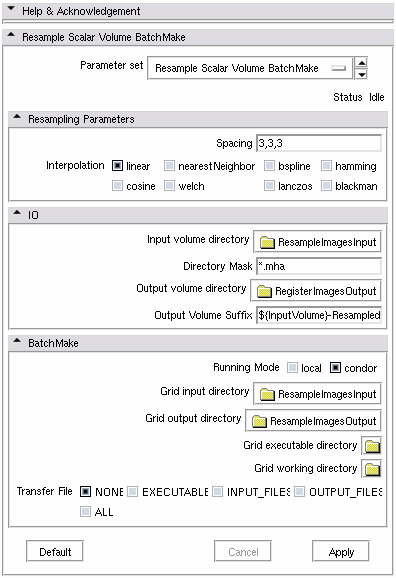
Input volume volume= directory where the input images are located
Directory mask = Images to resample must match the directory mask to be processed by the module
Output volume directory = directory where to save the output
Output volume suffix = name to give to the output files. To ensure uniqueness of the name, the BatchMake variable ${InputVolume} contains the name of each moving image.
Interpolation = <linear|nearestNeighbor|hamming|cosine|welch|lanczos|blackman> Sampling algorithm (linear, nearest neighbor or windowed sinc). Each window has a radius of 3; (default: linear) Spacing =Spacing along each dimension (0 means use input spacing) (default: 0,0,0)
Examples, Use Cases & Tutorials
- Use to resample volumes
In order to resample (linear interpolation) all the .mha volume files located in c:\images, and save the resampled version in c:\result where the file names are the input names suffixed by "-resampled", the following parameters are necessary. Input volume= c:\images Directory mask = *.mha Output volume directory = c:\result Output volume suffix = ${InputVolume}-resampled.mha Interpolation = linear Spacing = 2. 2. 2.
Quick Tour of Features and Use
If a condor grid is correctly configurated on the machine, the work can be shared with other computers.
Development
Dependencies
Known bugs
Don't use space character in paths Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Source Code: BatchMakeModule.cxx
XML Description: ResampleVolumeBatchMake.xml
Documentation: batchmake.org
More Information
Acknowledgment
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on the National Centers for Biomedical Computing can be obtained from National Centers for Biomedical Computing.