Modules:LabelStatistics-Documentation-3.6
Return to Slicer 3.6 Documentation
Module Name
Label Statistics
General Information
Module Type & Category
Type: Base
Category: Statistics
Authors, Collaborators & Contact
- Author: Katharina Quintus, BWH, Steve Pieper, Isomics
- Contact: pieper at bwh.harvard.edu
Module Description
Calculate image and volume properties using a label map and a gray scale image.
Usage
Examples, Use Cases & Tutorials
Quick Tour of Features and Use
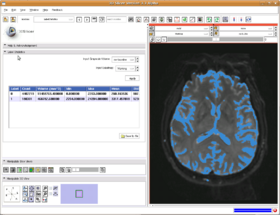
The module is fairly simple. Just select a background volume and a label volume (for example, as created using the Editor Module) and click Apply.
The volumes of the table show:
- Label there is one row for each unique value in the label volume
- Count is the number of pixels in the label volume that have this value
- Volume is the product of the pixel spacings (volume per pixel) times the Count
- Min, Max, Mean, StdDev Statistics on the grayscale pixel values at the locations that correspond to the label value.
Development
Dependencies
Other modules or packages that are required for this module's use.
Known bugs
Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Doxygen documentation:
More Information
Acknowledgment
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on the National Centers for Biomedical Computing can be obtained from National Centers for Biomedical Computing.