Documentation/Nightly/Modules/DTI ALPSIndex
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: DTI-ALPS Index | |||||
|
Module Description
This module offer the Diffusion Tensor Image Analysis Along the Perivascular Space (DTI-ALPS) scalar index, which is able to calculate the activity of the glymphatic system, which may be evaluated with diffusion images. Lower diffusivity along the perivascular space on DTI-APLS seems to reflect impairment of the glymphatic system. More details about the DTI-ALPS method is presented in the Taoka et al. 2017 original paper [1]
Use Cases
- Use Case 1: Obtain the DTI-ALPS index calculation from standard DTI image and propers anatomical labels (see details at Taoka et al. 2017 paper [2])
- The DTI-ALPS index is a scalar value that represents the activity of the glymphatic system, which may be evaluated with diffusion images. Lower diffusivity along the perivascular space on DTI-APLS seems to reflect impairment of the glymphatic system.
Usage Tips
- Tip 1: Call the DTI-ALPS calculation using GUI interface
- The DTI-ALPS index can be calculated using the direct GUI interface provided in the Slicer Module. See the User Interface section below.
- Tip 2: Call the DTI-ALPS calculation using command line Python script
- This Slicer Module provides a Python script to be called in the Slicer execution model, using command line. This can be useful when the user want to call a series of calculation without the use of GUI interface or by a server. See some ways to call the Python script
runDTIALPS.py:
- This Slicer Module provides a Python script to be called in the Slicer execution model, using command line. This can be useful when the user want to call a series of calculation without the use of GUI interface or by a server. See some ways to call the Python script
Assuming that Slicer is installed on Linux/Mac machines:
./Slicer --no-main-window --no-splash --python-script /path/to/module/runDTIALPS.py dti_volume.nrrd proj-label.nrrd assoc-label.nrrd
Assuming that Slicer is installed on Windows machines:
Slicer.exe --no-main-window --no-splash --python-script /path/to/module/runDTIALPS.py dti_volume.nrrd proj-label.nrrd assoc-label.nrrd
The runDTIALPS.py has some options that can be used:
1. --MNISpace: Informs whether the input DTI image is already in the MNI space (2 mm resolution). If yes, the input Proj/Assoc paths are changed for the standard MNI labels instead.
2. --verbose: Show more details thoughout the processing.
NOTE: When the --MNISpace is used, there is no need to provide the Proj/Assoc labels because the standard MNI labels are applied instead
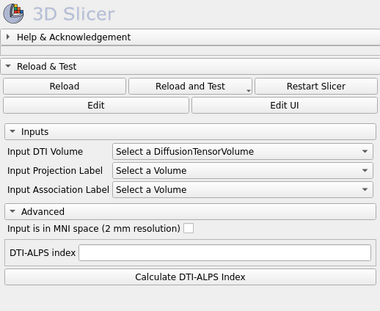
Panels and their use
Inputs:
- Input DTI Volume
- Input DTI volume. (Tip: Use Diffusion Tensor Estimation module to create an DTI image file)
- Input Projection Label
- The Projection label file that represents this brain region, as the same as described in the Taoka et al. 2017 paper.
- Input Association Label
- The Association label file that represents this brain region, as the same as described in the Taoka et al. 2017 paper.
- DTI-ALPS index
- The DTI-ALPS index value output
Advanced Parameters:
- Input is in MNI space (2 mm resolution)
- A checkbox that affirms whether the input DTI image is already in the MNI space (2 mm resolution brain template). If it is true, than the Projection and Association labels are loaded from the template folder automatically. This is useful to run in batch processing, when a group study is performed. Note: The DTI space to MNI standard in not applied, this must be done before calling the DTI-ALPS module.
Similar Modules
N/A
References
- Toshiaki Taoka, et al. "Evaluation of glymphatic system activity with the diffusion MR technique: diffusion tensor image analysis along the perivascular space (DTI-ALPS) in Alzheimer's disease cases", (2017). Jpn J Radiol, Apr;35(4):172-178. PMID: 28197821, DOI: 10.1007/s11604-017-0617-z
Information for Developers
| Section under construction. |