Documentation/4.6/Extensions/DTI-Reg
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: DTI-Reg | |||||||||
|
Module Description
DTI-Reg is an extension that performs pair-wise DTI registration, using scalar FA map to drive the registration.
Individual steps of the pair-wise registration pipeline are performed via external applications - some of them being 3D Slicer modules. Starting with two input DTI images, scalar FA maps are generated via dtiprocess. Registration is then performed between these FA maps, via BRAINSFit/BRAINSDemonWarp or ANTS -Advanced Normalization Tools-, which provide different registration schemes: rigid, affine, BSpline, diffeomorphic, logDemons. The final deformation is then applied to the source DTI image via ResampleDTIlogEuclidean.
Use Cases
Tutorials
A tutorial can be found here
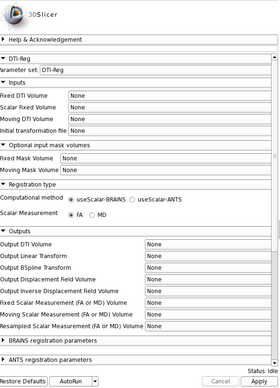
Panels and their use
Similar Modules
See ResampleDTIlogEuclidean and DTIAtlasBuilder.
References
Information for Developers
The source code of DTI-Reg is available on GitHub. It is published under an Apache License v2.0.