Documentation/4.4/Modules/fibertrack
|
For the latest Slicer documentation, visit the read-the-docs. |
This tool is deprecated!!! It is not part of DTIProcess anymore!
FiberTrack
FiberTrack
General Information
Module Type & Category
Type: CLI
Category: Diffusion.NIRALPipeline
Authors, Collaborators & Contact
Author: Casey Goodlett
Contributors:
Contact: name, email
Module Description
| Program title | FiberTrack |
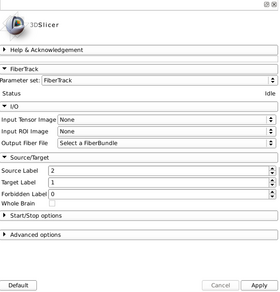
| Program description | \nThis program implements a simple streamline tractography method based on the principal eigenvector of the tensor field. A fourth order Runge-Kutta integration rule used to advance the streamlines.\nAs a first parameter you have to input the tensor field (with the --input_tensor_file option). Then the region of interest image file is set with the --input_roi_file. Next you want to set the output fiber file name after the --output_fiber_file option. \nYou can specify the label value in the input_roi_file with the --target_label, --source_label and --fobidden_label options. By default target label is 1, source label is 2 and forbidden label is 0. The source label is where the streamlines are seeded, the target label defines the voxels through which the fibers must pass by to be kept in the final fiber file and the forbidden label defines the voxels where the streamlines are stopped if they pass through it. There is also a --whole_brain option which, if enabled, consider both target and source labels of the roi image as target labels and all the voxels of the image are considered as sources.\nDuring the tractography, the --fa_min parameter is used as the minimum value needed at different voxel for the tracking to keep going along a streamline. The --step_size parameter is used for each iteration of the tracking algorithm and defines the length of each step. The --max_angle option defines the maximum angle allowed between two successive segments along the tracked fiber. |
| Program version | 1.1.0 |
| Program documentation-url | http://www.google.com/ |
Usage
Quick Tour of Features and Use
A list panels in the interface, their features, what they mean, and how to use them.
|
More Information
More information is available on the project webpage: http://www.nitrc.org/projects/dtiprocess
Acknowledgment
Hans Johnson(1,3,4); Kent Williams(1); (1=University of Iowa Department of Psychiatry, 3=University of Iowa Department of Biomedical Engineering, 4=University of Iowa Department of Electrical and Computer Engineering) provided conversions to make DTIProcess compatible with Slicer execution, and simplified the stand-alone build requirements by removing the dependancies on boost and a fortran compiler.