Documentation/4.4/Modules/ABC
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Author: Marcel Prastawa Acknowledgments: This project was supported by NIH grant U54 EB005149.
|
Module Description
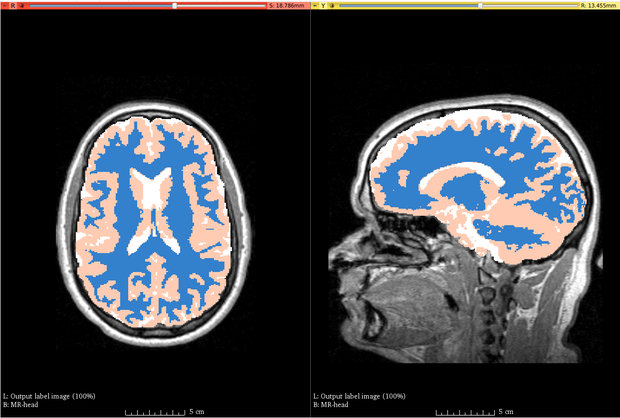
ABC (Atlas Based Classification) is a full segmentation pipeline designed and developed for healthy human brain MRIs, and can be adapted to other applications that involve multi-channel MR images and a pre-defined atlas. The processing pipeline includes image registration, filtering, inhomogeneity correction, and skull stripping.
Tutorials
The software is designed and tested for healthy brain MRI (pediatric - adult). Initiating the pipeline requires the specification of an atlas (which contains a template image and prior probability images) and at least one image modality for a subject. The module can accommodate up to five modalities, which need not be registered or skull-stripped beforehand. All images are assumed to have the correct metadata for spatial orientation, voxel spacing, and image origin.
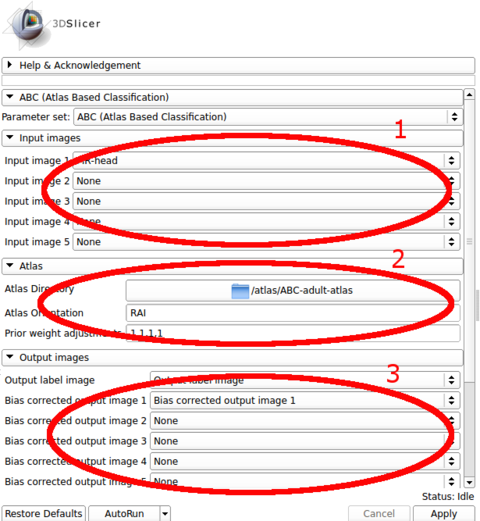
The input images are specified in the first panel. The choice of the first image in the list is crucial, as the outputs are generated in the space of the first image. All other image channels and the atlas are registered to the first image during processing.
The atlas can be specified through the second panel. The module assumes that it is stored in a directory where the template image (e.g., a T1 image) is specified as template.mha and that the prior images are stored in sequence (1.mha, 2.mha, ...). An example atlas is available through NITRC for adult brains: Adult Brain Atlas, which needs to be uncompressed and stored in a local path before running the module.
The output images are specified in the third panel. Users can create and specify the target volumes for the segmentation labels as well as the bias corrected images for each modality. The bias corrected output images do not need to be specified if they are of no interest.
Once all input parameters have been specified, execution can be initiated by clicking the Apply button.
The processing involved in the module can take up to five minutes for a typical MR image, using the default setting that was chosen to generate coarser results. Speed-ups are possible by increasing the number of threads (specified in the Speed panel). Results with higher accuracy can be obtained by increasing the maximum polynomial degree of the bias field (in the Advanced panel), increasing the number of pre-filtering iterations (in the Advanced panel), and switching to finer sampling for image registration (in the Speed panel).
Panels and their use
Parameters:
()
* ':
** ':
*** ':
List of parameters generated transforming this XML file using this XSL file. To update the URL of the XML file, edit this page.
Similar Modules
References
- Automated model-based tissue classification of MR images of the brain. K Van Leemput, F Maes, D Vandermeulen, P Suetens. Medical Imaging, IEEE Transactions on 18 (10), 897-908.
- Automated model-based bias field correction of MR images of the brain. K Van Leemput, F Maes, D Vandermeulen, P Suetens. Medical Imaging, IEEE Transactions on 18 (10), 885-896.
Information for Developers
| Section under construction. |
Source code can be found at http://www.nitrc.org/plugins/scmsvn/viewcvs.php/?root=abc