Documentation/4.4/Modules/Featurelet-Registration
Contents
Introduction and Acknowledgements
|
Extension: Slicer-Featurelets | |||||||
|
Module Description
The module "Featurelet-Registration" provides a featurelet-based, piecewise rigid, deformable image registration method. The so called "featurelets" are equally sized rectangular subvolumes of the moving image which are rigidly registered to rectangular search regions on the fixed image. The output is a deformed image and a deformation field.
The basic idea of the algorithm is from Söhn et al. (2008). Daniella Fabri, a former PhD Student at the general hospital of Vienna picked up this idea and developed an featurelet-based registration method which was intended to be used for image registration in the pelvic region. The algorithm was written in C++, but unfortunately without any GUI. Thus, it was not easy to use, especially for people without any programming skills.
The source code of Daniella Fabri was used to make a 3D-Slicer extension for Linux out of it. The extension consists of a single loadable module named “Featurelet-Registration”.
Use Cases
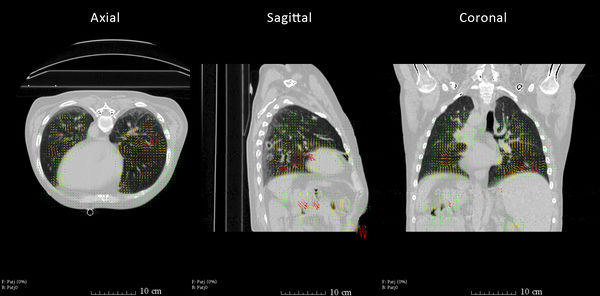 Example Registration: The axial, sagital and coronal view show the warped image of the registration of a maximum exhale to a maximum inhale state CT of a thorax. The corresponding output deformation field is visualized using the Transforms module. |
Tutorials
Panels and their use
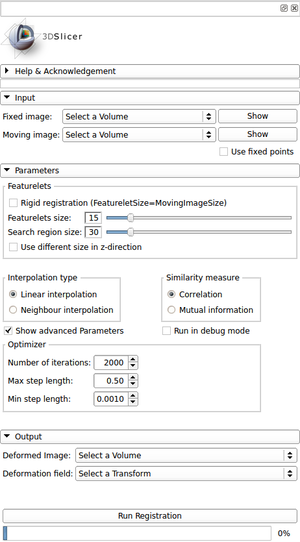
- Help & Acknowledgment:
- The Help-section provides the user with an exemplary workflow for image registration using the module Featurelet-Registration and information about other Slicer modules for pre- and post-processing of images and the registration output.
- The Acknowledgment-section provides information about the basic idea used for the module and its contributors.
- Input: In the Input-section the user can select the fixed and moving image intended to use for registration. Both images can be visualized immediately using the particular “Show”-button. In addition, the user can define fixed points in the image. A featurelet containing such a point is non moving.
- Parameters: In the Parameter-section both feturelet and search region size can be defined. Optionally a different size in the z-direction can be set for the case of large inter-slice spacings of an image. Regarding the registration process itself the user can select the type of interpolator (linear or nearest neighbor) and the metric used for measuring the voxel-based similarity (mutual information or normalized cross correlation). As advanced parameters optimizer settings can be changed and the registration process can be executed in debug mode. In this debug mode additional information about the registration process and its progress is shown in the terminal.
- Output: In the Output-section the user has to define the volume used for the deformed image as well as the one for the deformation field. By hitting the “Run Registration”-button the registration process starts and the progress is shown in the progress-bar below the button.
To facilitate usability several tooltips provide additional information for the user about the item being hovered over.
Similar Modules
References
- Söhn et al., “Model-independent, multimodality deformable image registration by local matching of anatomical features and minimization of elastic energy”, Medical Physics 35, 866–878, 2008.
Information for Developers
| Section under construction. |