Documentation/4.10/Modules/dtiestim
|
For the latest Slicer documentation, visit the read-the-docs. |
DTIEstim
DTIEstim
General Information
Module Type & Category
Type: CLI
Category: Diffusion.NIRALPipeline
Authors, Collaborators & Contact
Author: Casey Goodlett
Contributors:
Contact: name, email
Module Description
| Program title | DTIEstim |
| Program description | dtiestim is a tool that takes in a set of DWIs (with --dwi_image option) in nrrd format and estimates a tensor field out of it. The output tensor file name is specified with the --tensor_output option \nThere are several methods to estimate the tensors which you can specify with the option --method lls-wls-nls-ml . Here is a short description of the different methods: \n\tlls Linear least squares. Standard estimation technique that recovers the tensor parameters by multiplying the log of the normalized signal intensities by the pseudo-inverse of the gradient matrix. Default option.\n\twls Weighted least squares. This method is similar to the linear least squares method except that the gradient matrix is weighted by the original lls estimate. (See Salvador, R., Pena, A., Menon, D. K., Carpenter, T. A., Pickard, J. D., and Bullmore, E. T. Formal characterization and extension of the linearized diffusion tensor model. Human Brain Mapping 24, 2 (Feb. 2005), 144-155. for more information on this method). This method is recommended for most applications. The weight for each iteration can be specified with the --weight_iterations. It is not currently the default due to occasional matrix singularities.\n\tnls Non-linear least squares. This method does not take the log of the signal and requires an optimization based on levenberg-marquadt to optimize the parameters of the signal. The lls estimate is used as an initialization. For this method the step size can be specified with the --step option. \n\tml Maximum likelihood estimation. This method is experimental and is not currently recommended. For this ml method the sigma can be specified with the option --sigma and the step size can be specified with the --step option.\nYou can set a threshold (--threshold) to have the tensor estimated to only a subset of voxels. All the baseline voxel value higher than the threshold define the voxels where the tensors are computed. If not specified the threshold is calculated using an OTSU threshold on the baseline image.The masked generated by the -t option or by the otsu value can be saved with the --B0_mask_output option.\ndtiestim also can extract a few scalar images out of the DWI set of images: \n\tthe average baseline image (--B0) which is the average of all the B0s.\n\tthe IDWI (--idwi)which is the geometric mean of the diffusion images.\nYou can also load a mask if you want to compute the tensors only where the voxels are non-zero (--brain_mask) or a negative mask and the tensors will be estimated where the negative mask has zero values (--bad_region_mask) |
| Program version | 1.1.0 |
| Program documentation-url | http://www.nitrc.org/projects/dtiprocess |
Usage
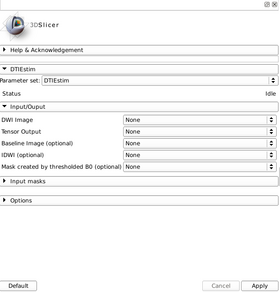
Quick Tour of Features and Use
A list panels in the interface, their features, what they mean, and how to use them.
|
More Information
More information is available on the project webpage: http://www.nitrc.org/projects/dtiprocess
Acknowledgment
Hans Johnson(1,3,4); Kent Williams(1); (1=University of Iowa Department of Psychiatry, 3=University of Iowa Department of Biomedical Engineering, 4=University of Iowa Department of Electrical and Computer Engineering) provided conversions to make DTIProcess compatible with Slicer execution, and simplified the stand-alone build requirements by removing the dependancies on boost and a fortran compiler.
References
Publications related to this module go here. Links to pdfs would be useful.