Slicer3:UIDesign:WorkingProblems:BCAnalysis:Dynamic
Back to Project Overview
Contents
Perfusion Analysis for Breast Cancer
Goal
Goal is to accommodate basic RECIST assessment for breast cancer (Response Evaluation Criteria In Solid Tumors). (RECIST is a set of published rules that define when cancer patients improve ("respond"), stay the same ("stable") or worsen ("progression") during treatments).
- Determine features that currently exist in Slicer
- Develop features that don't
- Provide a framework that knits features together in a comfortable workflow.
Perfusion Analysis Workflow
- Visualize the dataset dynamically
- Specify an ROI that includes tumor and apply to all timepoints Question: ROI or VOI? (either -- if slice, must be same slice)
- Plot contrast dilution curve
- show time to peak in tumor
- Specify an ROI that includes blood pool and apply to all timepoints Question: again, ROI or VOI?
- Plot contrast dilution curve for this (on same graph)
- show time to peak in blood pool
- show transit time (distance between peaks in each plot)
- save out timepoints ( Intensity(t) for each ROI -- include multiple tumors if desired)
First experiment
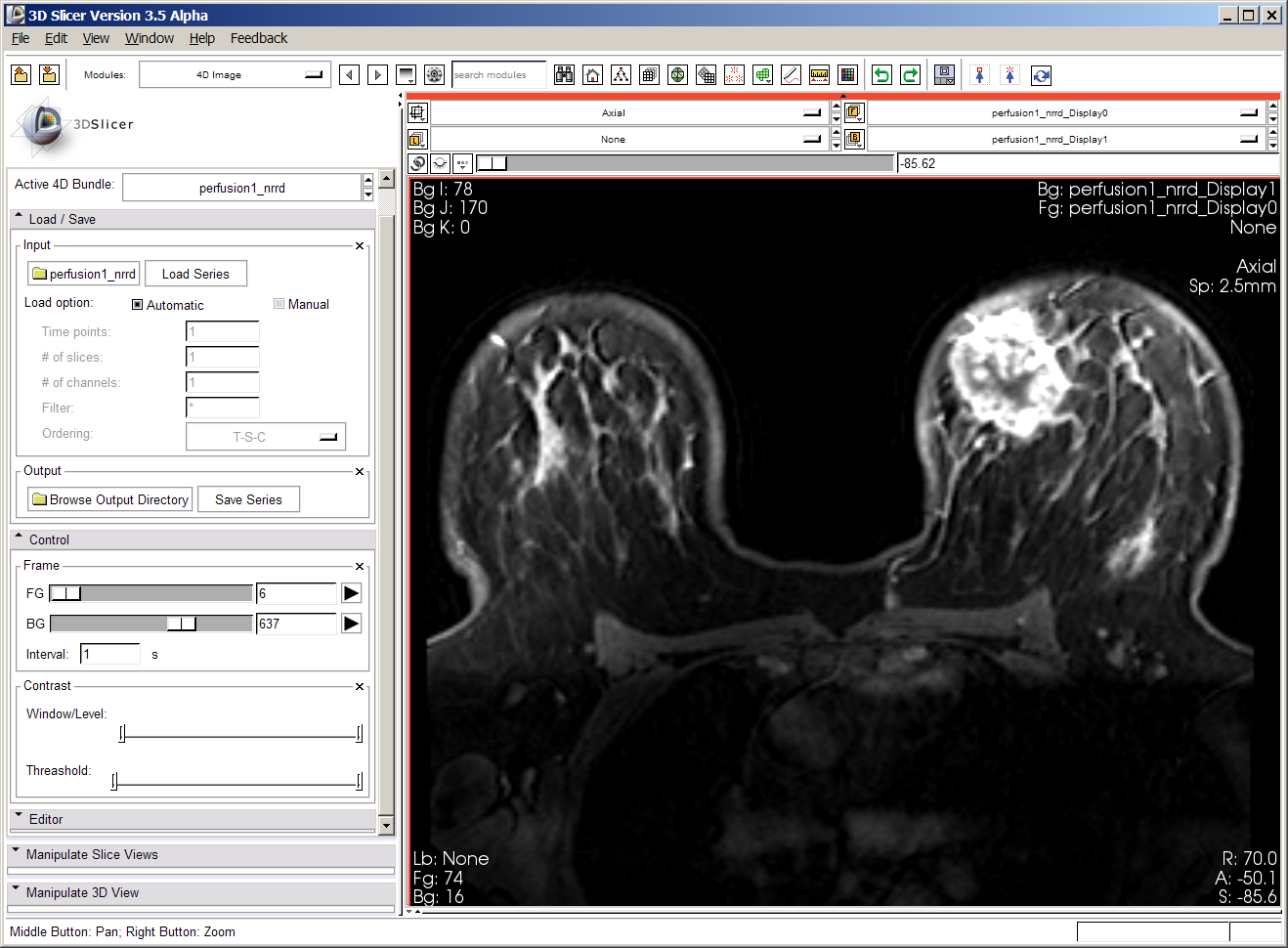
Below is a screenshot of first perfusion dataset. The dataset contains upward of 900 volumes. Loading the original DICOM images took a very long time; Junichi converted these images to nrrd format, but the loading still takes prohibitively long, and interacting is also very slow.
ideas:
- can we create a VOI and apply to each volume in the timeseries?
- Alternatively, for a first demo we can use a smaller dataset?
update:
From Jeff -- first dataset contains 15 timepoints, each 256x256x63 -- so we've found a bug in either:
- the timeseries loader in Slicer (probably in the thing that parses dicom header), or
- we have a malformed DICOM header.
This is causing Slicer to load 63x15 volumes containing a single 256x256 slice. I'll dig in to this today and see what's going on. Shouldn't be too hard to find. Junichi has given me a DCE dataset that loads properly for comparison.
The 4D Image analysis package is required for plotting -- need to expose this in Slicer3/Modules/CMakelists.txt file. Discussed with Junichi about possibility of having both 4DImage and 4DImageAnalysis modules exposed in the trunk for the demo; he sees no problem in this *without* the Scipy modeling part exposed. Too challenging now to build python/numpy/scipy by default.
- Exposed FourDImage and FourDAnalysis modules in Slicer3/Modules/CMakelists.txt
- Changed the code in vtkKWPlotGraph.cxx: replaced finite() function is replaced code that conforms to ISO C++/2003 so it would compile on win32
- Had to move pyconfig.h to Slicer3-lib/python-build/Include so FourDAnalysis module could compile (couldn't find this file, included by <Python.h> for some reason (maybe something wrong with my python install.