Difference between revisions of "Slicer3:BrainLab Integration"
From Slicer Wiki
| Line 14: | Line 14: | ||
== '''Step 1:''' Download and Install Software Packages == | == '''Step 1:''' Download and Install Software Packages == | ||
| + | We assume both BioImage Suite and Slicer3 will be installed on the same Windows XP computer. | ||
* Install BioImage Suite | * Install BioImage Suite | ||
** The latest binary of BioImage Suite (2.6 Release Candidate 1) can be installed from here: http://research.yale.edu/bioimagesuite/forum/index.php?topic=257.msg. This contains an up-to-date version of the OpenIGTLink interface. | ** The latest binary of BioImage Suite (2.6 Release Candidate 1) can be installed from here: http://research.yale.edu/bioimagesuite/forum/index.php?topic=257.msg. This contains an up-to-date version of the OpenIGTLink interface. | ||
Revision as of 18:22, 26 November 2008
Home < Slicer3:BrainLab IntegrationContents
Introduction
To better understand the integration of BrainLab and Slicer3, we first introduce the following terms:
- Slicer3 [[1]] is cross-platform end user application for analyzing and visualizing medical images. It contails collection of Open Source libraries for developing and deploying new image computing technologies.
- BrainLab [[2]] currently offers a set of integrated OR solutions, for instance neurosurgery, orthopedic, and RT/Oncology. The BrainLab component we're working with is VectorVision Cranial Navigation System.
- BioImage Suite [[3]] is an integrated image analysis software suite developed at Yale University.
- VectorVision Link (VVLink) is a custom designed client/server tool which extends functionality from the Visualization Toolkit. VV Link enables bi-directional data transfer such as image data sets, visualizations and tool positions in real time.
- OpenIGTLink is a simple network protocol intended for trackers, robots and other devices to send data to the main application.
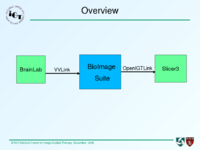
As you can see in Fig. 1, BioImage Suite communicates with BrainLab on VVLink and talk with Slicer3 through OpenIGTLink. The data flow is: BrainLab sends tracking data and images to BioImage Suite, which in turn relays them to Slicer3.
The Procedure
Please follow the following steps to use Slicer to do some research in DTI visualization during surgery. The procedure is being developed and tested on Windows XP. If you are using a different OS (e.g. Linux, or Mac), please contact us.
Step 1: Download and Install Software Packages
We assume both BioImage Suite and Slicer3 will be installed on the same Windows XP computer.
- Install BioImage Suite
- The latest binary of BioImage Suite (2.6 Release Candidate 1) can be installed from here: http://research.yale.edu/bioimagesuite/forum/index.php?topic=257.msg. This contains an up-to-date version of the OpenIGTLink interface.
- Install Slicer3
- Slicer3 binary download site: http://www.slicer.org/DownloadSlicer.php/Nightly
- Choose one Win32 installer and download it
- Run the installer to install Slicer3 on your Windows XP computer