Difference between revisions of "Modules:VMTKVesselEnhancement"
| Line 55: | Line 55: | ||
The following screencasts/tutorials show different Use Cases of the VMTKVesselEnhancement module. This module is always used as a pre-processing step and the actual segmentation is performed using the [[Modules:VMTKLevelSetSegmentation|VMTKLevelSetSegmentation]] module. | The following screencasts/tutorials show different Use Cases of the VMTKVesselEnhancement module. This module is always used as a pre-processing step and the actual segmentation is performed using the [[Modules:VMTKLevelSetSegmentation|VMTKLevelSetSegmentation]] module. | ||
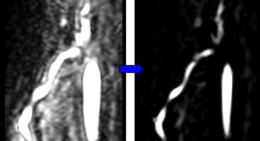
| − | In both tutorials the Sato Vesselness filter is used, which provides good results for highlighting vascular structures. | + | In both tutorials the ''Sato Vesselness'' filter is used, which provides good results for highlighting vascular structures. |
* '''Use Case #1''': Extraction of a Coronary Tree inside a cardiac blood-pool MRI by using only one fiducial point and Fast Marching | * '''Use Case #1''': Extraction of a Coronary Tree inside a cardiac blood-pool MRI by using only one fiducial point and Fast Marching | ||
Revision as of 05:58, 16 July 2009
Home < Modules:VMTKVesselEnhancementReturn to Slicer 3.4 Documentation
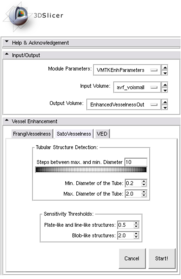
Module Name
VMTKVesselEnhancement
General Information
Module Type & Category
Type: Scripted Module
Category: Filtering, Extension
Authors, Collaborators & Contact
- Author: Daniel Haehn, University of Heidelberg
- Supervisor: Luca Antiga, Mario Negri Institute
- Contact: Daniel Haehn, haehn@bwh.harvard.edu
Module Description
This module provides the vessel enhancement filters of the Vascular Modeling Toolkit (http://www.vmtk.org) in 3D Slicer. These filters target the highlighting of line-like, plate-like and blob-like structures.
This work is part of the NA-MIC VMTK Collaboration.
Official project page: http://www.vmtk.org/Main/VmtkIn3DSlicer
Usage
Installation
This module depends on the VmtkSlicerModule: see this page for installation notes.
The VMTKVesselEnhancement module can be installed as a 3D Slicer Extension or manually.
- Installation as a 3D Slicer Extension
Inside the 3D Slicer extension system, the module is called VMTKVesselEnhancement.
- Manual Installation
1. To get the latest source code, perform the following SVN checkout command:
svn checkout https://www.nitrc.org/svn/slicervmtklvlst
2. A directory named VMTKVesselEnhancement is downloaded.
3. Copy this directory to Slicer3-build/lib/Slicer3/Modules/ of your local Slicer installation.
When the module was successfully installed, it is available within 3D Slicer's module selector inside the category Vascular Modeling Toolkit.
Examples, Use Cases & Tutorials
The following screencasts/tutorials show different Use Cases of the VMTKVesselEnhancement module. This module is always used as a pre-processing step and the actual segmentation is performed using the VMTKLevelSetSegmentation module.
In both tutorials the Sato Vesselness filter is used, which provides good results for highlighting vascular structures.
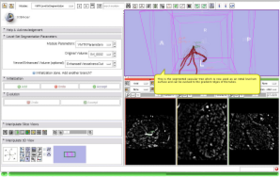
- Use Case #1: Extraction of a Coronary Tree inside a cardiac blood-pool MRI by using only one fiducial point and Fast Marching
 Watch the video.. |
This screencast shows the extraction of a coronary tree inside a cardiac blood-pool MRI by using a combination of the VMTKVesselEnhancement and the VMTKLevelSetSegmentation modules. Only one fiducial point is used as a source point for the Fast Marching algorithm. Watch the video here. |
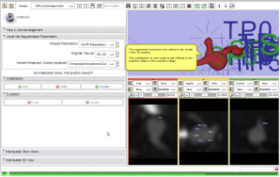
- Use Case #2: Segmentation of a Cerebral Aneurysm and its connecting vessels using Colliding Fronts and Fast Marching
 Watch the video.. |
The segmentation of a cerebral aneurysm and its connecting vessels is shown in this screencast. The VMTKVesselEnhancement module is used as a pre-processing step and the segmentation of the connecting vessels is performed on the vessel enhanced image with the Colliding Fronts algorithm using the VMTKLevelSetSegmentation module. The segmentation of the actual aneurysm is performed on the original image. How to switch between these images during the initialization stage easily is explained as well. Watch the video here. |
Quick Tour of Features and Use
Development
Dependencies
This module depends on the VMTK libraries which are provided in the VmtkSlicerModule. Therefore the VmtkSlicerModule has to be installed before the VMTKLevelSetSegmentation module can be used.
Known bugs & Usability issues
Follow this link to the VMTK in 3D Slicer bug tracker.
Source code & documentation
VMTKVesselEnhancement is a Python Scripted Module. It follows the conventions of the Model View Controller pattern of slicer modules. This implies the separation of logic and GUI.
The class SlicerVMTKVesselEnhancementGUI derives from ScriptedModuleGUI and saves the current user interface state to its own MRML node. The MRML nodes of the used volume and model data are additionally attached. This allows different instances of the vessel enhancement module to be run at the same time. The actual calls to the VMTK libraries are only performed in the class SlicerVMTKVesselEnhancementLogic.
Separate classes derived from the interface SlicerVMTKAdvancedPageSkeleton were created for each filtering method in order to ensure the possibility of maintenance and extension. The calls to the required methods like UpdateGUI() or UpdateMRML() get forwarded to each individual sub-class to support the modular design and enable code encapsulation.
The complete source code is available at a NITRC SVN repository.
More Information
Acknowledgment
This work was funded by a grant of the Thomas-Gessmann Foundation part of the Founder Federation for German Science.
References
- Piccinelli M, Veneziani A, Steinman DA, Remuzzi A, Antiga L (2009) A framework for geometric analysis of vascular structures: applications to cerebral aneurysms. IEEE Trans Med Imaging. In press.
- Antiga L, Piccinelli M, Botti L, Ene-Iordache B, Remuzzi A and Steinman DA. An image-based modeling framework for patient-specific computational hemodynamics. Medical and Biological Engineering and Computing, 46: 1097-1112, Nov 2008.