Difference between revisions of "Modules:LabelDiameterEstimation-Documentation-3.5"
| Line 29: | Line 29: | ||
This module was originally designed to calculate the dimensions of tumor, following the RECIST/WHO guidelines. | This module was originally designed to calculate the dimensions of tumor, following the RECIST/WHO guidelines. | ||
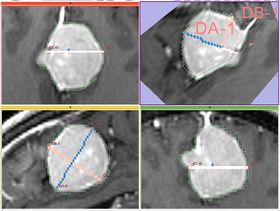
| − | * '''Implementation details''' The module implements the following algorithm for calculating the dimensions of the label. First, the area of the label slice is computed in all the slices of the image that are passing through the label. This is done for the three orthogonal directions of the image. The largest area slice is found. ''The largest diameter (DA)'' is estimated by finding the two most distant points on the contour of the label cross-section. ''The second diameter (DB)'' is found by finding the two most distant points on the contour that lie on the line perpendicular to the first diameter. ''The third diameter (DC)'' is estimated by calculating the two points of intersection between the line perpendicular to the plane formed by ''DA'' and ''DB'' | + | * '''Implementation details''' The module implements the following algorithm for calculating the dimensions of the label. First, the area of the label slice is computed in all the slices of the image that are passing through the label. This is done for the three orthogonal directions of the image. The largest area slice is found. ''The largest diameter (DA)'' is estimated by finding the two most distant points on the contour of the label cross-section. ''The second diameter (DB)'' is found by finding the two most distant points on the contour that lie on the line perpendicular to the first diameter. ''The third diameter (DC)'' is estimated by calculating the two points of intersection between the line perpendicular to the plane formed by ''DA'' and ''DB''passing through the point of their intersection, and the contour of the analyzed label. |
===Quick Tour of Features and Use=== | ===Quick Tour of Features and Use=== | ||
Revision as of 01:11, 16 November 2009
Home < Modules:LabelDiameterEstimation-Documentation-3.5Return to Slicer 3.5 Documentation
Label Diameter Estimation
LabelDiameterEstimation
General Information
Module Type & Category
Type: CLI
Category: Statistics
Authors, Collaborators & Contact
- Andriy Fedorov, BWH
- Ron Kikinis, BWH
- Contact: Andriy Fedorov, fedorov at bwh
Module Description
Given a binary label, this module estimates the largest diameter of the label, its largest in-plane perpendicular diameter, and the third diameter that is perpendicular to the plane of the first two.
Usage
Examples, Use Cases & Tutorials
This module was originally designed to calculate the dimensions of tumor, following the RECIST/WHO guidelines.
- Implementation details The module implements the following algorithm for calculating the dimensions of the label. First, the area of the label slice is computed in all the slices of the image that are passing through the label. This is done for the three orthogonal directions of the image. The largest area slice is found. The largest diameter (DA) is estimated by finding the two most distant points on the contour of the label cross-section. The second diameter (DB) is found by finding the two most distant points on the contour that lie on the line perpendicular to the first diameter. The third diameter (DC) is estimated by calculating the two points of intersection between the line perpendicular to the plane formed by DA and DBpassing through the point of their intersection, and the contour of the analyzed label.
Quick Tour of Features and Use
List all the panels in your interface, their features, what they mean, and how to use them. For instance:
- Input panel:
- Parameters panel:
- Output panel:
- Viewing panel:
Development
Dependencies
Known bugs
Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Source code can accessed here
Links to documentation generated by doxygen.
More Information
Acknowledgment
Supported by Brain Science Foundation.