Difference between revisions of "EMSegmenter-Tasks:Template"
| Line 4: | Line 4: | ||
=Description= | =Description= | ||
| − | -- Data Assumption | + | -- Data Assumption <BR> |
-- description of pipeline with citations to algorithms used | -- description of pipeline with citations to algorithms used | ||
Revision as of 21:50, 8 December 2010
Home < EMSegmenter-Tasks:TemplateReturn to EMSegmenter Task Overview Page
Contents
This wiki template can be used for new tasks
Description
-- Data Assumption
-- description of pipeline with citations to algorithms used
Anatomical Tree
- root
- background (BG)
- air (AIR)
- intracranial cavity (ICC)
- white matter (WM)
- background (BG)
Atlas
- how did you construct the atlas ?
-- what did you do for skull stripping or any other preprocessing
-- What did you do for group registration
-- how did you generate the segmentations
-- how many set of images did you use
-- where did you get the data from
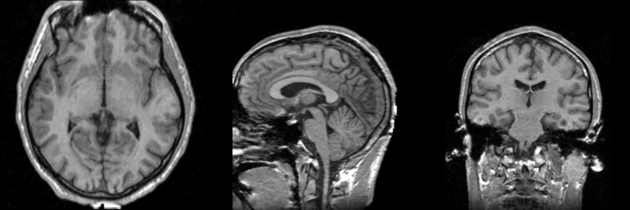
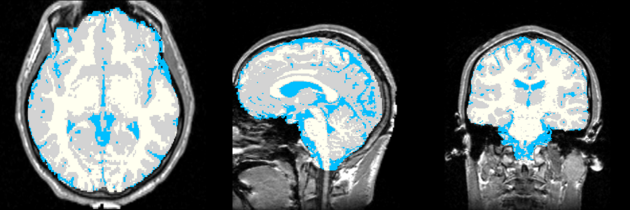
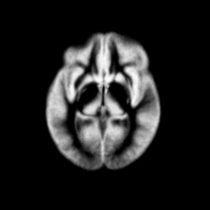
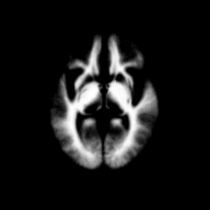
Atlas was generated based on 82 scans and corresponding segmentations provided by Psychiatry Neuroimaging Laboratory, BWH. We registered the scans to a preselected template via Warfield et al. 2001.
Image Dimension = 256 x 256 x 124
Image Spacing = 0.9375 x 0.9375 x 1.5
|
|
|
|
| Template (T1) | CSF | GM | WM |
Result
Acknowledgment
The construction of the pipeline was supported by funding from NIH NCRR 2P41RR013218 Supplement.
Citations
- Pohl K, Bouix S, Nakamura M, Rohlfing T, McCarley R, Kikinis R, Grimson W, Shenton M, Wells W. A Hierarchical Algorithm for MR Brain Image Parcellation. IEEE Transactions on Medical Imaging. 2007 Sept;26(9):1201-1212.