EMSegmenter-Tasks
Return to EMSegmenter Overview Page
Contents
What is a task?
Most clinicians are using the EMSegmenter to segment specific parts of the human body. Those segmentations depend heavily on
the used input data. The signal level in the input data varies with the used image modalities (e.g. MRI, CT, ...) and with each anatomical structure.
The EMSegmenter can take advantage of some knowledge about the signal level of those anatomical structures in the different image modalities.
Technically, each tasks consist of a .mrml file and a .tcl file.
The .mrml file stores the anatomical properties (mean values, covariance values) in a user defined tree structure.
The .tcl script is used to perform some pre-processing on the input data.
Visit the following web page to create your own task.
Segmentation Task Library
Existing Tasks in Slicer 3.6.3
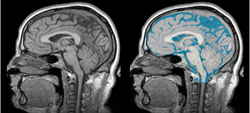 Task 01: MRI Human Brain for non-skull stripped T1 scans
Task 01: MRI Human Brain for non-skull stripped T1 scans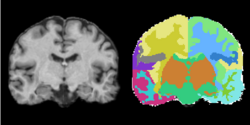 Task 02: MRI Human Brain Parcellation for skull stripped T1 scans
Task 02: MRI Human Brain Parcellation for skull stripped T1 scans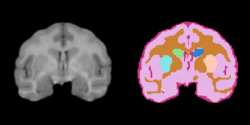 Task 03: Non-Human Primate for skull stripped T1 scans
Task 03: Non-Human Primate for skull stripped T1 scans
Tasks in development for Slicer 3.6.4
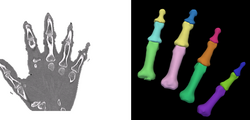 Task 04: CT Hand Bone in development
Task 04: CT Hand Bone in development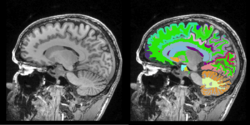 Task 05: MRI Human Brain Full Parcellation for non-skull stripped T1 scans
Task 05: MRI Human Brain Full Parcellation for non-skull stripped T1 scans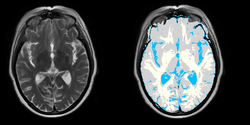 Task 06: MRI Human Brain with high in-plane resolution for non-skull stripped T1 scans
Task 06: MRI Human Brain with high in-plane resolution for non-skull stripped T1 scans
EMSegmenter use cases
Quantitative assessment using MPRAGE and Flair images
- Collaborator: Tammie Benzinger , Washington University School of Medicine
- Quick description: Quantitative assessment using MPRAGE and Flair images
- Image specification: T2 MPRAGE
- Used Task: MRI Human Brain (with skull stripping enabled)