Difference between revisions of "EMSegmenter-Tasks"
Belhachemi (talk | contribs) |
|||
| (89 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | [[EMSegmenter-Overview|Return to EMSegmenter Overview Page]] | |
| + | |||
| + | |||
| + | =What is a task?= | ||
Most clinicians are using the EMSegmenter to segment specific parts of the human body. Those segmentations depend heavily on | Most clinicians are using the EMSegmenter to segment specific parts of the human body. Those segmentations depend heavily on | ||
| Line 10: | Line 13: | ||
The .tcl script is used to perform some pre-processing on the input data.<br> | The .tcl script is used to perform some pre-processing on the input data.<br> | ||
<br> | <br> | ||
| + | Visit the following web page to create your own [[EMSegmenter-CreateTask| task]] or we can [[ EMSegmenter-UseCaseAdvertisement | help ]] you . | ||
| − | + | =Segmentation Task Library= | |
| − | =Existing Tasks | + | ==Existing Tasks in Slicer 3.6.3== |
| − | |||
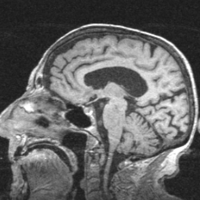
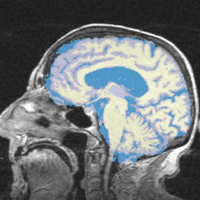
*[[Image:MRI-Human-Brain_BeforeAfter.png|250px]] '''Task 01: [[EMSegmenter-Tasks:MRI-Human-Brain|MRI Human Brain ''' for non-skull stripped T1 scans]] | *[[Image:MRI-Human-Brain_BeforeAfter.png|250px]] '''Task 01: [[EMSegmenter-Tasks:MRI-Human-Brain|MRI Human Brain ''' for non-skull stripped T1 scans]] | ||
*[[Image:MRIHumanBrainParcellation.png|250px]] '''Task 02: [[EMSegmenter-Tasks:MRI-Human-Brain-Parcellation|MRI Human Brain Parcellation ''' for skull stripped T1 scans]] | *[[Image:MRIHumanBrainParcellation.png|250px]] '''Task 02: [[EMSegmenter-Tasks:MRI-Human-Brain-Parcellation|MRI Human Brain Parcellation ''' for skull stripped T1 scans]] | ||
| + | *[[Image:NHP.png|250px]] '''Task 03: [[EMSegmenter-Tasks:Non-Human-Primate|Non-Human Primate ''' for skull stripped T1 scans]] | ||
| − | =Tasks | + | ==Tasks for Slicer 3.6.4== |
| − | *[[Image: | + | * [[Image:EMS Result Hand subject2.png|250px]] '''Task 04: [[EMSegmenter-Tasks:CT-Hand-Bone|CT Hand Bone ''']] |
| − | * | + | * [[Image:MRIHumanBrainFullParcellation_before_after.png|250px]] '''Task 05: [[EMSegmenter-Tasks:MRI-Human-Brain-Full-Parcellation|MRI Human Brain Full Parcellation''' for non-skull stripped T1 scans]] |
| + | * [[Image:MRIHumanBrainHIPR beforeafter.png|250px]] '''Task 06: [[EMSegmenter-Tasks:MRI-Human-Brain-HIPR|MRI Human Brain with high in-plane resolution''' for non-skull stripped T1 scans]] | ||
| + | * [[Image:EMS_Hemisphere_before_after.png|250px]]'''Task 07: [[EMSegmenter-Tasks:MRI-Human-Brain-Hemisphere|MRI Human Brain Hemisphere''' for non-skull stripped T1 scans]] | ||
| + | |||
| + | ==Tasks in development== | ||
| + | * [[Image:EMS HumanEye Before After.png|250px]]'''Task 08: [[EMSegmenter-Tasks:Human-Eye|Human Eye''']] | ||
| + | * [[Image:EMS MRIHumanBrainExp Before After.png|250px]] '''Task 09: [[EMSegmenter-Tasks:MRI-Human-Brain-Experimental|MRI Human Brain Experimental''' with skull stripping]] | ||
| + | * [[Image:EMSegment-CardioA1-grey-small.jpg|125px]][[Image:EMSegment CardioA1small.jpg|125px]] '''Task 10: [[EMSegmenter-Tasks:cineMARI|cine MRI''' ]] | ||
| + | * [[Image:EMS LesionsExp BeforeAfter.png|250px]] '''Task 11: [[EMSegmenter-Tasks:LesionExp|White matter lesion segmentation''' ]] | ||
| + | * '''Task 12: [[EMSegmenter-Tasks:EMS_Spine|Bone segmentation''' ]] | ||
| + | ==External Command Lines (Under Development) == | ||
| + | * [[Image:GLISTR Flair.png|125px]][[Image:GLISTR Seg.png|125px]]'''Task 12: [https://www.rad.upenn.edu/sbia/projects/glistr.html GLISTR: Automatic Glioma Segmentation'''] <BR> The software was developed and is distributed by SBIA. SBIA will soon release an interface for GLISTR to interact with Slicer | ||
| − | =Task | + | =Additional Use Cases= |
| − | + | ==Quantitative assessment using MPRAGE and Flair images== | |
| + | * Collaborator: Tammie Benzinger , Washington University School of Medicine | ||
| + | * Short description: Quantitative assessment using MPRAGE and Flair images | ||
| + | * Image specification: Dimension: 256x256x160 , Spacing 1x1x1, T2 MPRAGE | ||
| + | * Used Task: [[EMSegmenter-Tasks:MRI-Human-Brain-Experimental|'''MRI Human Brain Experimental''' (with skull stripping)]] | ||
| + | <gallery perrow=2: widths=200px : heights=200px> | ||
| + | Image:EMS_Quantitative_assessment_orig.png | ||
| + | Image:EMS_Quantitative_assessment.png | ||
| + | </gallery> | ||
| − | + | ==Brain Atrophy in Aging Patients== | |
| − | + | * Collaborator: David Tate, PI (Brigham and Women's Hospital), Daniel McCaffrey (Brigham and Women's Hospital), Alex Zaitsev (Brigham and Women's Hospital/Surgical Planning Laboratory) and Jewish Hospital | |
| − | + | * Short description: Evaluate Brain Atrophy in aging subjects (NOTE: the segmentation was done for old scans from 90s. No other tools could segment CSF.) | |
| − | + | * Image specification: NA Tesla, Scanner: NA, Psuedo T1, Axial Scans, TR/TI/TE=NA, pixel_xsize = 0.937500, pixel_ysize = 0.937500, fov = 240.000000, aspect = 0.000000, thick = 5.000000 | |
| − | + | * Used Task: [[EMSegmenter-Tasks:MRI-Human-Brain|'''MRI Human Brain''']] | |
| − | + | <gallery perrow=2: widths=200px : heights=200px> | |
| − | + | Image:EMS_AgingStudy_DTate_Project_3-6-2.png | |
| − | + | Image:EMS_AgingStudy_PseudoT1_OldMIRs_JewishHospEMS_manualTrain_60x.png | |
| − | + | </gallery> | |
| + | ==Brain Atrophy in HIV Patients== | ||
| + | * Collaborator: David Tate, PI (Brigham and Women's Hospital), Troy Russel (Brigham and Women's Hospital), Alex Zaitsev (Brigham and Women's Hospital/Surgical Planning Laboratory) and National HIV Consortium | ||
| + | * Short description: Evaluate Brain Atrophy in HIV Patients | ||
| + | * Image specification: 3 Tesla, Scanner: NA, MPRAGE, Sagittal Scans, TR/TI/TE=NA, pixel_xsize = 0.9375, pixel_ysize = 0.9375, fov = NA, aspect = NA, space = 1.5 | ||
| + | * Used Task: [[EMSegmenter-Tasks:MRI-Human-Brain|'''MRI Human Brain''']] | ||
| + | <gallery perrow=2: widths=425px : heights=200px> | ||
| + | Image:EMS_HIVStudy_Segmentation.png | ||
| + | Image:EMS_HIVStudy_Segmentation_Auto.png | ||
| + | </gallery> | ||
| − | * | + | ==Pediatric MS Study== |
| + | * Collaborator: Tanuja Chitnis PI (Brigham and Women's Hospital/Multiple Sclerosis Center), Alexander Zaitsev (Brigham and Women's Hospital/Surgical Planning Laboratory) and Massachusetts General Hospital | ||
| + | * Short description: Evaluate brain atrophy for pediatric MS Patients | ||
| + | * Image specification: 3 Tesla, Scanner: SIGNA HDx / GE MEDICAL SYSTEMS, 3D MPRAGE, Sagital Scans, TR/TI/TE=24/0/7 ms, pixel_xsize = 0.976600, pixel_ysize = 0.976600, fov = 250.009598, aspect = 1.535941, thick = 1.500000, space = 0.000000 | ||
| + | * Used Task: [[EMSegmenter-Tasks:MRI-Human-Brain|'''MRI Human Brain''']] | ||
| + | <gallery perrow=1: widths=630px : heights=210px> | ||
| + | Image:EMS_PED_MS_Study_Segmentation.png | ||
| + | </gallery> | ||
| − | * | + | ==Social Cognition Study== |
| + | * Collaborator: Collaborator: Andrea Mike, MD (Pecs University, Hungary), Charles Guttmann, MD, Alexander Zaitsev (Brigham and Women's Hospital) | ||
| + | * Short description: Evaluate brain atrophy for elderly MS patients. | ||
| + | * Number of processed cases: 50 | ||
| + | * Image specification: 3 Tesla, Scanner: Magnetom TIM Trio, Siemens, 3D MPRAGE, Slice thickness: 1.2mm, Spacing 1.2 1 1 (mm) | ||
| + | * Used Task: [[EMSegmenter-Tasks:MRI-Human-Brain|'''MRI Human Brain''']] | ||
| + | <gallery perrow=1: widths=630px : heights=200px> | ||
| + | Image:EMS_SocialCognitionLESIONS_MANUALLY_SEGMENTED_PECS_S006_Seg.png | ||
| + | </gallery> | ||
| − | + | ==Physiological Brain Activity Longitudinal Study== | |
| − | * | + | * Collaborator: Laura Horky PI (Brigham and Women's Hospital(BWH)), Jon Hainer (BWH), Wendy Plesniak (BWH), Alexander Zaitsev (BWH), Ron Kikinis (BWH) and BWH Division of Nuclear Medicine |
| − | + | * Short description: Evaluate glycolitical brain activity in CSF and White Matter for the patients undergoing chemical therapy courses. | |
| − | + | * Image specification: 1.5 Tesla ,Scanner: SIGNA EXCITE / GE MEDICAL SYSTEMS, T1, Axial Scans, TR/TI/TE=500/0/21 ms, pixel_xsize = 0.781251, pixel_ysize = 0.781244, fov = 0.000000, aspect = 7.679990, thick = 5.000000, space = 1.000000 | |
| − | + | * Used Task: [[EMSegmenter-Tasks:MRI-Human-Brain-Hemisphere|'''MRI Human Brain Hemisphere''']] | |
| − | + | <gallery perrow=3: widths=250px : heights=200px> | |
| − | + | Image:EmSegment PETCT 1.png | |
| − | + | Image:EmSegment PETCT 2.png | |
| − | + | Image:EmSegment PETCT 3.png | Result on test data | |
| − | * | + | </gallery> |
| − | |||
| − | |||
| − | |||
| − | * | ||
| − | |||
| − | |||
| − | |||
| − | * | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | <gallery perrow=3: widths=250px : heights=200px> | ||
| + | Image:EMSegment PETCT Physiology-1.png | ||
| + | Image:EMSegment PETCT Physiology-2.png | ||
| + | Image:EMSegment PETCT Physiology-3.png | Result on patient data | ||
| + | </gallery> | ||
| − | The | + | ==Comprehensive Longitudinal Investigation of Multiple Sclerosis at Brigham and Women’s Hospital (CLIMB) Study== |
| − | + | * [http://www.partnersmscenter.org/index.php?id=217&mn=9 CLIMB Study Web Page] | |
| + | * Collaborator: Svetlana Egorova MD/PhD (Brigham and Women's Hospital(BWH)), Alexander Zaitsev PhD (BWH) | ||
| + | * Short description: Provide Basic EM Segmentation for estimation of brain parenchymal fraction (BPF) . The processing is being performed for the cases with failing PD TDS+ segmentation. The T1-type scans are segmented and registered to PD/T2 images. The requests are being submitted over the Web GUI. | ||
| + | * Image specification: 1.5 Tesla ,Scanner: Varies among participants. | ||
| + | * Used Task: [[EMSegmenter-Tasks:MRI-Human-Brain-Hemisphere|'''MRI Human Brain Hemisphere''']] | ||
Latest revision as of 00:42, 6 August 2011
Home < EMSegmenter-TasksReturn to EMSegmenter Overview Page
Contents
- 1 What is a task?
- 2 Segmentation Task Library
- 3 Additional Use Cases
- 3.1 Quantitative assessment using MPRAGE and Flair images
- 3.2 Brain Atrophy in Aging Patients
- 3.3 Brain Atrophy in HIV Patients
- 3.4 Pediatric MS Study
- 3.5 Social Cognition Study
- 3.6 Physiological Brain Activity Longitudinal Study
- 3.7 Comprehensive Longitudinal Investigation of Multiple Sclerosis at Brigham and Women’s Hospital (CLIMB) Study
What is a task?
Most clinicians are using the EMSegmenter to segment specific parts of the human body. Those segmentations depend heavily on
the used input data. The signal level in the input data varies with the used image modalities (e.g. MRI, CT, ...) and with each anatomical structure.
The EMSegmenter can take advantage of some knowledge about the signal level of those anatomical structures in the different image modalities.
Technically, each tasks consist of a .mrml file and a .tcl file.
The .mrml file stores the anatomical properties (mean values, covariance values) in a user defined tree structure.
The .tcl script is used to perform some pre-processing on the input data.
Visit the following web page to create your own task or we can help you .
Segmentation Task Library
Existing Tasks in Slicer 3.6.3
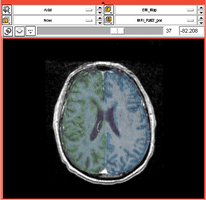
 Task 01: MRI Human Brain for non-skull stripped T1 scans
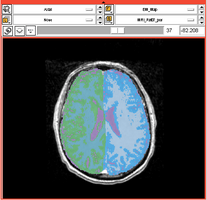
Task 01: MRI Human Brain for non-skull stripped T1 scans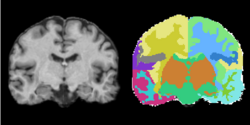 Task 02: MRI Human Brain Parcellation for skull stripped T1 scans
Task 02: MRI Human Brain Parcellation for skull stripped T1 scans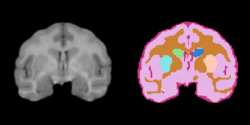 Task 03: Non-Human Primate for skull stripped T1 scans
Task 03: Non-Human Primate for skull stripped T1 scans
Tasks for Slicer 3.6.4
 Task 04: CT Hand Bone
Task 04: CT Hand Bone 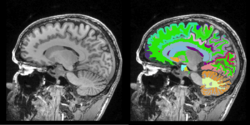 Task 05: MRI Human Brain Full Parcellation for non-skull stripped T1 scans
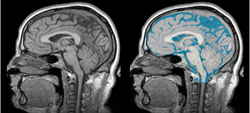
Task 05: MRI Human Brain Full Parcellation for non-skull stripped T1 scans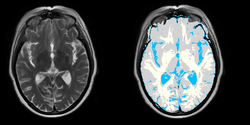 Task 06: MRI Human Brain with high in-plane resolution for non-skull stripped T1 scans
Task 06: MRI Human Brain with high in-plane resolution for non-skull stripped T1 scans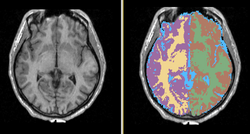 Task 07: MRI Human Brain Hemisphere for non-skull stripped T1 scans
Task 07: MRI Human Brain Hemisphere for non-skull stripped T1 scans
Tasks in development
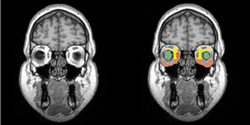 Task 08: Human Eye
Task 08: Human Eye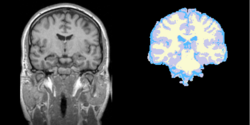 Task 09: MRI Human Brain Experimental with skull stripping
Task 09: MRI Human Brain Experimental with skull stripping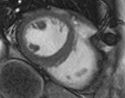
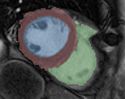 Task 10: cine MRI
Task 10: cine MRI 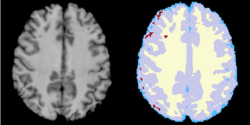 Task 11: White matter lesion segmentation
Task 11: White matter lesion segmentation - Task 12: Bone segmentation
External Command Lines (Under Development)
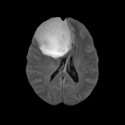
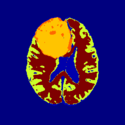 Task 12: GLISTR: Automatic Glioma Segmentation
Task 12: GLISTR: Automatic Glioma Segmentation
The software was developed and is distributed by SBIA. SBIA will soon release an interface for GLISTR to interact with Slicer
Additional Use Cases
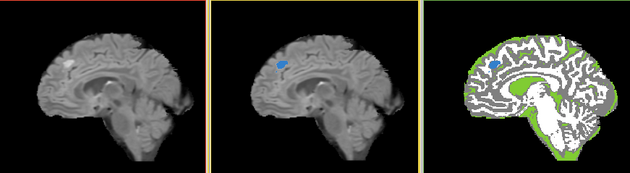
Quantitative assessment using MPRAGE and Flair images
- Collaborator: Tammie Benzinger , Washington University School of Medicine
- Short description: Quantitative assessment using MPRAGE and Flair images
- Image specification: Dimension: 256x256x160 , Spacing 1x1x1, T2 MPRAGE
- Used Task: MRI Human Brain Experimental (with skull stripping)
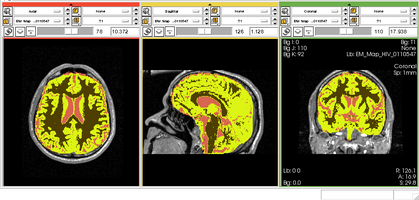
Brain Atrophy in Aging Patients
- Collaborator: David Tate, PI (Brigham and Women's Hospital), Daniel McCaffrey (Brigham and Women's Hospital), Alex Zaitsev (Brigham and Women's Hospital/Surgical Planning Laboratory) and Jewish Hospital
- Short description: Evaluate Brain Atrophy in aging subjects (NOTE: the segmentation was done for old scans from 90s. No other tools could segment CSF.)
- Image specification: NA Tesla, Scanner: NA, Psuedo T1, Axial Scans, TR/TI/TE=NA, pixel_xsize = 0.937500, pixel_ysize = 0.937500, fov = 240.000000, aspect = 0.000000, thick = 5.000000
- Used Task: MRI Human Brain
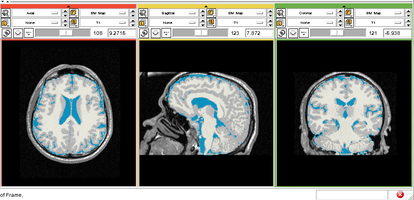
Brain Atrophy in HIV Patients
- Collaborator: David Tate, PI (Brigham and Women's Hospital), Troy Russel (Brigham and Women's Hospital), Alex Zaitsev (Brigham and Women's Hospital/Surgical Planning Laboratory) and National HIV Consortium
- Short description: Evaluate Brain Atrophy in HIV Patients
- Image specification: 3 Tesla, Scanner: NA, MPRAGE, Sagittal Scans, TR/TI/TE=NA, pixel_xsize = 0.9375, pixel_ysize = 0.9375, fov = NA, aspect = NA, space = 1.5
- Used Task: MRI Human Brain
Pediatric MS Study
- Collaborator: Tanuja Chitnis PI (Brigham and Women's Hospital/Multiple Sclerosis Center), Alexander Zaitsev (Brigham and Women's Hospital/Surgical Planning Laboratory) and Massachusetts General Hospital
- Short description: Evaluate brain atrophy for pediatric MS Patients
- Image specification: 3 Tesla, Scanner: SIGNA HDx / GE MEDICAL SYSTEMS, 3D MPRAGE, Sagital Scans, TR/TI/TE=24/0/7 ms, pixel_xsize = 0.976600, pixel_ysize = 0.976600, fov = 250.009598, aspect = 1.535941, thick = 1.500000, space = 0.000000
- Used Task: MRI Human Brain
Social Cognition Study
- Collaborator: Collaborator: Andrea Mike, MD (Pecs University, Hungary), Charles Guttmann, MD, Alexander Zaitsev (Brigham and Women's Hospital)
- Short description: Evaluate brain atrophy for elderly MS patients.
- Number of processed cases: 50
- Image specification: 3 Tesla, Scanner: Magnetom TIM Trio, Siemens, 3D MPRAGE, Slice thickness: 1.2mm, Spacing 1.2 1 1 (mm)
- Used Task: MRI Human Brain
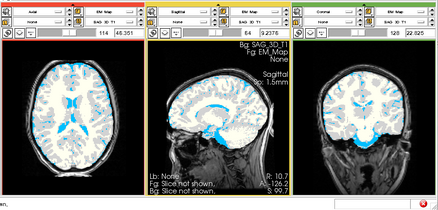
Physiological Brain Activity Longitudinal Study
- Collaborator: Laura Horky PI (Brigham and Women's Hospital(BWH)), Jon Hainer (BWH), Wendy Plesniak (BWH), Alexander Zaitsev (BWH), Ron Kikinis (BWH) and BWH Division of Nuclear Medicine
- Short description: Evaluate glycolitical brain activity in CSF and White Matter for the patients undergoing chemical therapy courses.
- Image specification: 1.5 Tesla ,Scanner: SIGNA EXCITE / GE MEDICAL SYSTEMS, T1, Axial Scans, TR/TI/TE=500/0/21 ms, pixel_xsize = 0.781251, pixel_ysize = 0.781244, fov = 0.000000, aspect = 7.679990, thick = 5.000000, space = 1.000000
- Used Task: MRI Human Brain Hemisphere
Comprehensive Longitudinal Investigation of Multiple Sclerosis at Brigham and Women’s Hospital (CLIMB) Study
- CLIMB Study Web Page
- Collaborator: Svetlana Egorova MD/PhD (Brigham and Women's Hospital(BWH)), Alexander Zaitsev PhD (BWH)
- Short description: Provide Basic EM Segmentation for estimation of brain parenchymal fraction (BPF) . The processing is being performed for the cases with failing PD TDS+ segmentation. The T1-type scans are segmented and registered to PD/T2 images. The requests are being submitted over the Web GUI.
- Image specification: 1.5 Tesla ,Scanner: Varies among participants.
- Used Task: MRI Human Brain Hemisphere