Difference between revisions of "EMSegmenter-Tasks"
Belhachemi (talk | contribs) |
Belhachemi (talk | contribs) (→Legend) |
||
| Line 101: | Line 101: | ||
WM: white matter <br> | WM: white matter <br> | ||
CSF: cerebrospinal fluid <br> | CSF: cerebrospinal fluid <br> | ||
| − | cau: caudate | + | cau: caudate <br> |
| − | put: putamen | + | put: putamen <br> |
| − | hip: hippocampus | + | hip: hippocampus <br> |
| − | ICC: Intracranial Cavity | + | ICC: Intracranial Cavity <br> |
WGC: <br> | WGC: <br> | ||
lSGM: <br> | lSGM: <br> | ||
Revision as of 22:38, 23 November 2010
Home < EMSegmenter-TasksUnder construction
What is a task?
Most clinicians are using the EMSegmenter to segment specific parts of the human body. Those segmentations depend heavily on
the used input data. The signal level in the input data varies with the used image modalities (e.g. MRI, CT, ...) and with each anatomical structure.
The EMSegmenter can take advantage of some knowledge about the signal level of those anatomical structures in the different image modalities.
Technically, each tasks consist of a .mrml file and a .tcl file.
The .mrml file stores the anatomical properties (mean values, covariance values) in a user defined tree structure.
The .tcl script is used to perform some pre-processing on the input data.
Existing Tasks
There are currently 2 tasks defined in Slicer 3.6.2
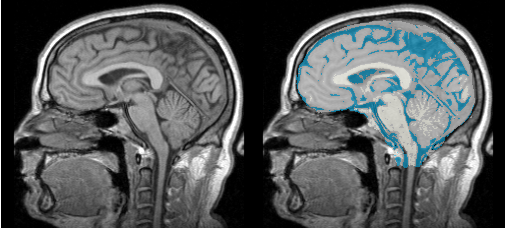
MRI Human Brain
Description
MRI Human Head pipeline for a finer-grained parcellation based on data provided by
Anatomical Tree
- ROOT
- BG
- AIR
- Skull
- ICC
- WM
- GM
- CSF
- BG
Result
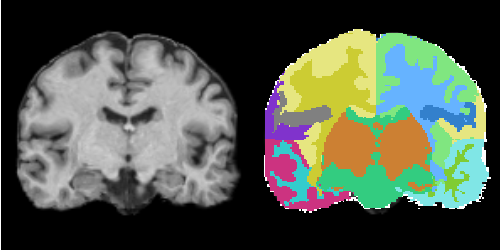
MRI Human Brain Parcellation
Description
MRI Human Head pipeline for a finer-grained parcellation based on data provided by
Anatomical Tree
Legend:
BG: background
LT: left temporal
GM: grey matter
WM: white matter
CSF: cerebrospinal fluid
Tree:
- Root
- BG
- GM
- LTGM
- LTGM1
- LTGM2
- LTGM3
- LTGM4
- RTGM
- RTGM1
- RTGM2
- RTGM3
- RTGM4
- SUBGM
- LTGM
- WM
- LTWM
- LTWM1
- LTWM2
- LTWM3
- LTWM4
- RTWM
- RTWM1
- RTWM2
- RTWM3
- RTWM4
- SUBWM
- LTWM
- CSF
Result
Collaborators
Padmapriya Srinivasan and Sylvain Bouix (PNL-BWH)
Tasks in development
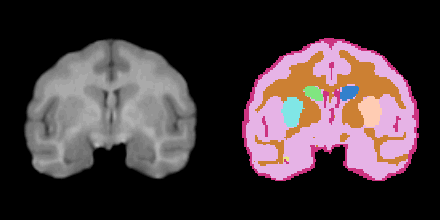
Non-Human Primate
Legend
BG: background
LT: left temporal
GM: grey matter
WM: white matter
CSF: cerebrospinal fluid
cau: caudate
put: putamen
hip: hippocampus
ICC: Intracranial Cavity
WGC:
lSGM:
rSGM:
BS:
CB:
Anatomical Tree
- Root
- Head
- ICC
- WGC
- GM
- lSGM
- lcau
- lput
- lhip
- rSGM
- rcau
- rput
- rhip
- GMleaf
- lSGM
- WM
- CSF
- GM
- BS
- CB
- WGC
- rest
- ICC
- Head
Result
Collaborators
Andriy Fedorov (BWH)
CT Hand Bone
Collaborators
TBD
Task creation process - for developer
The goal is to create a task with the name 'Hello World'
After following the instructions below the new task will consist of following files:
./Slicer3/Modules/EMSegment/Tasks/Hello-World.mrml ./Slicer3/Modules/EMSegment/Tasks/Hello-World.tcl ./Slicer3/Modules/EMSegment/Tasks/Hello-World/<atlas_file_1> ./Slicer3/Modules/EMSegment/Tasks/Hello-World/<atlas_file_2> [...] ./Slicer3/Modules/EMSegment/Tasks/Hello-World/<atlas_file_i> [...] ./Slicer3/Modules/EMSegment/Tasks/Hello-World/<atlas_file_N>
- Create a new directory ./Slicer3/Modules/EMSegment/Tasks/Hello-World
- Copy the atlas files for air, GM, T1 into this directory.
- Start Slicer
- Switch to the EMSegmenter module
- Create new task
- Name: Hello World
- Pre-processing: None
- Basically follow our EMSegmenter advanced tutorial, but create a simple tree
- load ./Slicer3/Modules/EMSegment/Testing/TestData/MiscVolumeData/MRIHumanBrain_T1_aligned.nrrd
- Add a channel, call it T1, assign MRIHumanBrain_T1_aligned
- Create a simple anatomical tree (air and GM)
- Load atlas files only from ./Slicer3/Modules/EMSegment/Tasks/Hello-World/
- generate mean and covariance values
- specify node weights
- At step 9/9. click on 'Create Template File'
- Save the file under ./Slicer3/Modules/EMSegment/Tasks/Hello-World.mrml
- The target node and some other unneeded nodes will be removed from the mrml scene and written to disk.
- Close Slicer
- Copy our standard tcl file ./Slicer3/Modules/EMSegment/Tasks/Template.tcl to ./Slicer3/Modules/EMSegment/Tasks/Hello-World.tcl
- Adjust this tcl file to include your pre-processing pipeline.
- edit ./Slicer3/Modules/EMSegment/Tasks/Hello-World.tcl to specify your own pre-processing
- edit ./Slicer3/Modules/EMSegment/Tasks/Hello-World.mrml to use your own pre-processing
- - <EMS TclTaskFilename="GenericTask.tcl" ></EMS>
- + <EMS TclTaskFilename="Hello-World.tcl" ></EMS>
- touch ../Slicer3/Modules/EMSegment/CMakeLists.txt and do a 'make' to copy the new files into the binary directory