Difference between revisions of "Documentation:Nightly:Registration:RegistrationLibrary:RegLib C42"
| (One intermediate revision by the same user not shown) | |||
| Line 12: | Line 12: | ||
|} | |} | ||
| − | == Modules | + | == Modules used == |
| + | *[[Documentation/Nightly/Modules/BRAINSFit| ''General Registration (BRAINS)'']] | ||
| + | |||
| + | == Download (from NAMIC MIDAS) == | ||
| + | <small>''Why 2 sets of files? The "input data" mrb includes only the unregistered data to try the method yourself from start to finish. The full dataset includes intermediate files and results (transforms, resampled images etc.). If you use the full dataset we recommend to choose different names for the images/results you create yourself to distinguish the old data from the new one you generated yourself. ''</small> | ||
| + | *[http://slicer.kitware.com/midas3/download/?items=119557 '''RegLib_C42.mrb'''] <br><small>(input data only, Slicer mrb file. 12 MB) </small><br> | ||
| + | *[http://slicer.kitware.com/midas3/download/?items=119556 '''RegLib_C42_full.mrb''']<br><small>(input data + results, Slicer mrb file. 36 MB). </small> | ||
== Description == | == Description == | ||
This is a typical case of change assessment. | This is a typical case of change assessment. | ||
| − | |||
| − | |||
| − | |||
=== Keywords === | === Keywords === | ||
MRI, brain, head, intra-subject, T1, autism, change assessment, pediatric MRI | MRI, brain, head, intra-subject, T1, autism, change assessment, pediatric MRI | ||
| − | + | == Procedure == | |
| − | + | #'''Affine Registration''': open the [[Documentation/Nightly/Modules/BRAINSFit| ''General Registration (BRAINS)'']] module. | |
| − | + | ##''Fixed Image Volume'': "Autism_T1_4yrs" | |
| − | # | + | ##''Moving Volume'': "Autism_T1_2yrs" |
| − | ## | + | ##Registration phases: check boxes for ''Rigid" ,"Scale", "Affine" |
| − | ## | + | ##''Slicer Linear Transform'': select "create new transform", rename to "Xf1_Affine" or similar |
| − | ## | + | ##leave rest at defaults. Click ''Apply'' |
| − | ## | + | ##registration should take ~ 10 secs. |
| − | ### | + | ##use fade slider to verify alignment; compare with result snapshots shown below. Alignment will not be perfect but should be better than before. |
| − | ##Output | + | ##note: you can also change the colormaps for the fixed and moving volumes to better judge the alignment: go to the [[Documentation/Nightly/Modules/Volumes| ''Volumes'']] module and in the ''Display'' tab, select "green" and "magenta" as the respective colormaps for the two volumes (vhf, vhm) |
| − | ## | + | #'''Nonrigid Registration''': open the [[Documentation/Nightly/Modules/BRAINSFit| ''General Registration (BRAINS)'']]. |
| − | ## | + | ##''Fixed/Moving Image Volume'': as above |
| − | + | ##Registration phases: uncheck boxes for rigid, scale and affine and check box for ''BSpline'' | |
| − | ##Registration Parameters: | + | ##Output: Slicer Linear transform: set to None |
| − | + | ##Output: Slicer BSpline transform: create new, rename to "Xf2_BSpline" or similar | |
| − | + | ##Output Image Volume: create new, rename to "Autism_T1_2yrs_Xf2"; ''Pixel Type'': "short" | |
| + | ##Registration Parameters: increase ''Number Of Samples'' to 300,000; ''Number of Grid Subdivisions'': 5,5,5 | ||
##Leave all other settings at default | ##Leave all other settings at default | ||
| − | ##click | + | ##click apply |
| − | |||
| + | == Registration Results== | ||
[[Image:RegLib_C42_unregistered.gif|300px]] baseline and follow-up before registration (click to enlarge) <br> | [[Image:RegLib_C42_unregistered.gif|300px]] baseline and follow-up before registration (click to enlarge) <br> | ||
[[Image:RegLib_C42_affine.gif|300px]] after affine alignment. Note the residual shape differences (click to enlarge) <br> | [[Image:RegLib_C42_affine.gif|300px]] after affine alignment. Note the residual shape differences (click to enlarge) <br> | ||
Latest revision as of 19:02, 1 October 2013
Home < Documentation:Nightly:Registration:RegistrationLibrary:RegLib C42Contents
Slicer Registration Library Case #42:
Intra-subject Brain MRI: serial assessment of brain development in autism
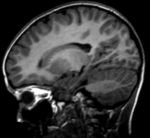
Input
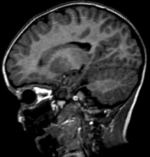
|
|
|
| fixed image/target | moving image |
Modules used
Download (from NAMIC MIDAS)
Why 2 sets of files? The "input data" mrb includes only the unregistered data to try the method yourself from start to finish. The full dataset includes intermediate files and results (transforms, resampled images etc.). If you use the full dataset we recommend to choose different names for the images/results you create yourself to distinguish the old data from the new one you generated yourself.
- RegLib_C42.mrb
(input data only, Slicer mrb file. 12 MB) - RegLib_C42_full.mrb
(input data + results, Slicer mrb file. 36 MB).
Description
This is a typical case of change assessment.
Keywords
MRI, brain, head, intra-subject, T1, autism, change assessment, pediatric MRI
Procedure
- Affine Registration: open the General Registration (BRAINS) module.
- Fixed Image Volume: "Autism_T1_4yrs"
- Moving Volume: "Autism_T1_2yrs"
- Registration phases: check boxes for Rigid" ,"Scale", "Affine"
- Slicer Linear Transform: select "create new transform", rename to "Xf1_Affine" or similar
- leave rest at defaults. Click Apply
- registration should take ~ 10 secs.
- use fade slider to verify alignment; compare with result snapshots shown below. Alignment will not be perfect but should be better than before.
- note: you can also change the colormaps for the fixed and moving volumes to better judge the alignment: go to the Volumes module and in the Display tab, select "green" and "magenta" as the respective colormaps for the two volumes (vhf, vhm)
- Nonrigid Registration: open the General Registration (BRAINS).
- Fixed/Moving Image Volume: as above
- Registration phases: uncheck boxes for rigid, scale and affine and check box for BSpline
- Output: Slicer Linear transform: set to None
- Output: Slicer BSpline transform: create new, rename to "Xf2_BSpline" or similar
- Output Image Volume: create new, rename to "Autism_T1_2yrs_Xf2"; Pixel Type: "short"
- Registration Parameters: increase Number Of Samples to 300,000; Number of Grid Subdivisions: 5,5,5
- Leave all other settings at default
- click apply
Registration Results
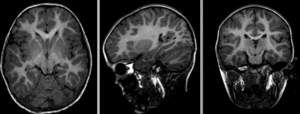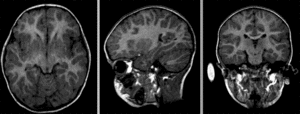 baseline and follow-up before registration (click to enlarge)
baseline and follow-up before registration (click to enlarge)
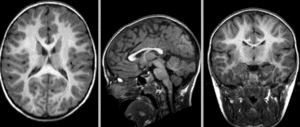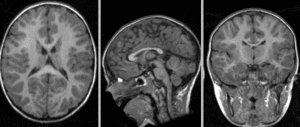 after affine alignment. Note the residual shape differences (click to enlarge)
after affine alignment. Note the residual shape differences (click to enlarge)
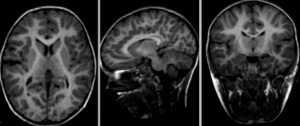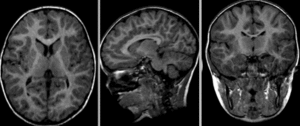 after nonrigid BSpline registration (click to enlarge)
after nonrigid BSpline registration (click to enlarge)
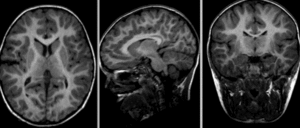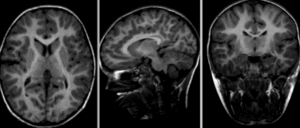 | visualization of the nonrigid deformation component only
| visualization of the nonrigid deformation component only