Difference between revisions of "Documentation:Nightly:Registration:RegistrationLibrary:RegLib C01"
From Slicer Wiki
| Line 92: | Line 92: | ||
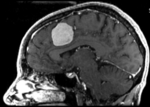
|[[Image:RegLib_C01_unregistered.png|300px]] ||baseline and follow-up before registration (click to enlarge) | |[[Image:RegLib_C01_unregistered.png|300px]] ||baseline and follow-up before registration (click to enlarge) | ||
|- | |- | ||
| − | |[[Image:RegLib_C01_rigid|300px]]||after rigid alignment. Note the residual shape differences (click to enlarge) | + | |[[Image:RegLib_C01_rigid.png|300px]]||after rigid alignment. Note the residual shape differences (click to enlarge) |
|- | |- | ||
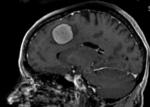
|[[Image:RegLib_C01_affine.png|300px]]||after affine registration (click to enlarge) | |[[Image:RegLib_C01_affine.png|300px]]||after affine registration (click to enlarge) | ||
Revision as of 20:45, 19 August 2013
Home < Documentation:Nightly:Registration:RegistrationLibrary:RegLib C01Contents
Slicer Registration Library Case #1: Follow-up MRI of Brain Tumor
Input
| fixed image | moving image |
Objective / Background
Goal is to align the follow-up image with the reference/baseline scan to assess changes.
Modules used
Download (from NAMIC MIDAS)
Keywords
MRI, brain, head, intra-subject, meningioma
Video Screencasts
- Movie/screencast showing generating a registration mask to exclude the tumor
- Movie/screencast showing rigid and affine registration
Procedure
- Rigid Registration: open the General Registration (BRAINS) module
- Input Images: Fixed Image Volume: MRMeningioma_1
- 'Input Images: Moving Image Volume: MRMeningioma_2
- Output Settings:
- Slicer Linear Transform (create new transform, rename to: "Xf1_Rigid")
- Output Image Volume none
- Registration Phases: select/check Rigid
- Initialization Phases: none
- Main Parameters:
- Leave all other settings at default
- click: Apply; runtime < 10 sec
- Building Registration Masks: We have now generate two masks that exclude the tumor region from contributing to the registration.
- open the Editor module
- As Master Volume, select the MR_1_baseline image
- click on the threshold tool and lower the threshold until the entire volume is selected (about a threshold of 5). Then click Apply.
- select the Brush tool. Increase the radius so that the brush completely covers the tumor (about 18mm).
- set the paint color to 0 (background).
- place the brush on an axial slice over the tumor. Use a left click to paint and the arrow keys to switch to the next slice. Repeat for all slices showing the tumor.
- save the mask as "MR_1_baseline_mask.nrrd"
- Now we generate a mask for MR_2. We can repeat the above or we can generate a mask by sending the first one through the inverse of the rigid transform we already computed.
- Go to the Transforms module.
- Active Transform: select the "Xf1_Rigid" transform we created above
- scroll down and click on the Invert button. This inverts the transform, which now points from MR_1 to MR_2. So now we can use it to generate a mask for MR_2 from the mask of MR_1. We must not forget to undo the invert afterwards!
- Go to the BRAINSResample module
- Image To Warp: select MR_1_baseline_mask
- Reference Image: select MR_2_followup
- Outputs:OutputImage: create & rename new, rename to "MR_2_followup_mask"
- Pixel Type: select uint or uchar
- Transform File: select "Xf1_Rigid"
- Interpolation Mode: select Nearest Neighbor
- Click Apply.
- Return to the Transforms module and invert the "Xf1_Rigid" transform again to return it to its original state.
- Go to the Volumes module
- Active Volume: select the new "MR_2_followup_mask"
- scroll down and check the 'labelmap checkbox.
- save the new mask.
- we now have two masks for use in the affine registration
- Affine registration: We have now increase the DOF to 12 in the form of an affine transform that also corrects scale and allows shear.
- Input Images: Fixed Image Volume: MR_1_baseline
- 'Input Images: Moving Image Volume: MR_2_followup
- Output Settings:
- Slicer Linear Transform (create new transform, rename to: "Xf2_Affine")
- Output Image Volume none
- Registration Phases: uncheck Rigid, select "Rigid+Scale" and "Affine"
- Initialization Transform: select "Xf1_Rigid"
- Initialization Transform Mode: off
- Main Parameters:
- Number Of Samples: increase to 300,000
- Masking Option: select "ROI" button
- Masking input fixed: select "MR_1_baseline_mask"
- Masking input moving: select "MR_2_followup_mask"
- click: Apply
- Verify
- place "MR_1_baseline" in the background and "MR_2_followup" into the foreground.
- place fade slider halfway
- Go to the Data module
- Drag "MR_2_followup" into the "Xf2_Affine" transform node, if it isn't there already.
Registration Results
 |
baseline and follow-up before registration (click to enlarge) |
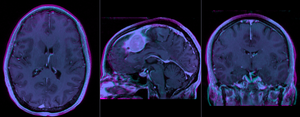 |
after rigid alignment. Note the residual shape differences (click to enlarge) |
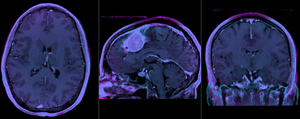 |
after affine registration (click to enlarge) |
Discussion: Key Strategies
- The affine registration seeks to address residual distortions that are typical for longitudinal MRI. However to prevent the tumor (and putative tumor change) from affecting the registration, we mask out the tumor so that it does not contribute to the registration. The assumption is thus that aside from the tumor we expect the two serial scans to be matching exactly, i.e. any differences in size or shape is considered an artifact we wish to remove by registration.