Difference between revisions of "Documentation/Nightly/Extensions/BrainTissuesExtension"
Acsenrafilho (talk | contribs) (Added the BrainTissuesExtension wikipage) |
Acsenrafilho (talk | contribs) m |
||
| (11 intermediate revisions by the same user not shown) | |||
| Line 27: | Line 27: | ||
[[Image:BrainTissuesExtension-logo.png|left]] | [[Image:BrainTissuesExtension-logo.png|left]] | ||
| − | + | Brain tissue segmentation plays an important role in many different image processing steps, which offers a possibility to study different neurodegenerative diseases or also in more specific tasks such as in brain structural quantitative morphological parameters such as brain atrophy. For this reason, brain tissue segmentation procedures have been intensively studied in recent years. This extension aims to offer a fast, robust and simple solutions for brain tissue segmentation, in which specific segmentation tasks are separated into different modules. See the modules list below to find the most appropriate tool for your study. | |
| + | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|Modules}} | {{documentation/{{documentation/version}}/extension-section|Modules}} | ||
| − | *[[Documentation/{{documentation/version}}/Modules/|... | + | |
| + | * '''General purpose segmentation method''': [[Documentation/{{documentation/version}}/Modules/BrainStructuresSegmenter|Brain Structures Segmenter]] | ||
| + | * '''Clustering classification based on k-Means algorithm''': [[Documentation/{{documentation/version}}/Modules/BasicBrainTissues|Basic Brain Tissues (k-Means)]] | ||
| + | * '''Multiple logistic classification algorithm''': [[Documentation/{{documentation/version}}/Modules/BrainLogisticSegmentation|Brain Logistic Segmentation (BLS)]] | ||
| + | |||
| + | '''NOTE''': The [[Documentation/{{documentation/version}}/Modules/BrainStructuresSegmenter|Brain Structures Segmenter]] is the main module for a general brain segmentation approach, which uses a predefined image processing pipeline in order to result in a smoothed tissue mask. See the module's documentation for more details. | ||
| + | |||
| + | Each segmentation approach is related to a specific algorithm, which is explained in more details in the pulications placed in References section. Please, cite the proper algorithm if any of these methods were used in a scientific study. | ||
| + | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Use Cases}} | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
Most frequently used for these scenarios: | Most frequently used for these scenarios: | ||
| − | * Use Case 1: a | + | * Use Case 1: Tissue classification |
| − | ** | + | ** There are several image quantitative approaches that are applied only in a certain tissue type (for instance, cortical thickness) in which a previous brain segmentation could be needed. |
| − | <gallery widths=" | + | * Use Case 2: Restrict brain regions for image quantitative analysis |
| − | Image: | + | ** A white matter mask could be used for specific measurements in some brain diseases, for example, in Multiple Sclerosis it could be useful to help lesion detection algorithms. |
| − | Image: | + | |
| − | Image: | + | <gallery widths="300px" heights="300px" perrow="3"> |
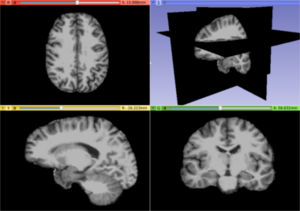
| − | Image: | + | Image:T1_fullBrain.png|A T1 weighted MRI image illustrating a usual brain |
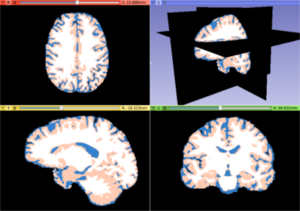
| − | Image: | + | Image:T1_tissues.png|White matter, gray matter and CSF tissues segmented from the previous MRI image |
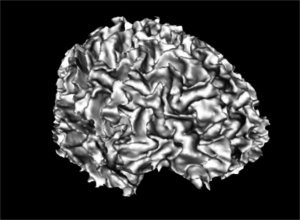
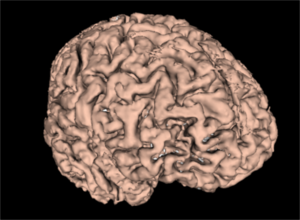
| + | Image:WM_3DReconstruction.png|A white matter mask 3D reconstruction | ||
| + | Image:GM_3DReconstruction.png|A gray matter mask 3D reconstruction | ||
| + | Image:WM_GM_3DReconstruction.png|A merged white and gray matter masks 3D reconstruction | ||
</gallery> | </gallery> | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|Similar Extensions}} | {{documentation/{{documentation/version}}/extension-section|Similar Extensions}} | ||
| − | + | [https://www.slicer.org/wiki/Modules:EMSegmenter-3.6 EM Segmenter] | |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|References}} | {{documentation/{{documentation/version}}/extension-section|References}} | ||
| − | * paper | + | |
| + | * [k-Means] | ||
| + | * [BLS paper] | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 59: | Line 73: | ||
Repositories: | Repositories: | ||
| − | * Source code: [https://github.com/CSIM-Toolkits/ GitHub repository] | + | * Source code: [https://github.com/CSIM-Toolkits/BrainTissuesExtension GitHub repository] |
| − | * Issue tracker: [https://github.com/CSIM-Toolkits/ open issues and enhancement requests] | + | * Issue tracker: [https://github.com/CSIM-Toolkits/BrainTissuesExtension/issues open issues and enhancement requests] |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-footer}} | {{documentation/{{documentation/version}}/extension-footer}} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 16:30, 30 October 2017
Home < Documentation < Nightly < Extensions < BrainTissuesExtension
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work was partially funded by CAPES and CNPq, a Brazillian Agencies. Information on CAPES can be obtained on the CAPES website and CNPq website. | |||||||||
|
Extension Description
Brain tissue segmentation plays an important role in many different image processing steps, which offers a possibility to study different neurodegenerative diseases or also in more specific tasks such as in brain structural quantitative morphological parameters such as brain atrophy. For this reason, brain tissue segmentation procedures have been intensively studied in recent years. This extension aims to offer a fast, robust and simple solutions for brain tissue segmentation, in which specific segmentation tasks are separated into different modules. See the modules list below to find the most appropriate tool for your study.
Modules
- General purpose segmentation method: Brain Structures Segmenter
- Clustering classification based on k-Means algorithm: Basic Brain Tissues (k-Means)
- Multiple logistic classification algorithm: Brain Logistic Segmentation (BLS)
NOTE: The Brain Structures Segmenter is the main module for a general brain segmentation approach, which uses a predefined image processing pipeline in order to result in a smoothed tissue mask. See the module's documentation for more details.
Each segmentation approach is related to a specific algorithm, which is explained in more details in the pulications placed in References section. Please, cite the proper algorithm if any of these methods were used in a scientific study.
Use Cases
Most frequently used for these scenarios:
- Use Case 1: Tissue classification
- There are several image quantitative approaches that are applied only in a certain tissue type (for instance, cortical thickness) in which a previous brain segmentation could be needed.
- Use Case 2: Restrict brain regions for image quantitative analysis
- A white matter mask could be used for specific measurements in some brain diseases, for example, in Multiple Sclerosis it could be useful to help lesion detection algorithms.
Similar Extensions
References
- [k-Means]
- [BLS paper]
Information for Developers
| Section under construction. |
Repositories:
- Source code: GitHub repository
- Issue tracker: open issues and enhancement requests