Difference between revisions of "Documentation/4.3/Announcements"
m (→Summary) |
|||
| (14 intermediate revisions by 6 users not shown) | |||
| Line 11: | Line 11: | ||
{|align="center" border="1" style="text-align:center; font-size:120%; border-spacing: 0; padding: 0px;" cellpadding="10" | {|align="center" border="1" style="text-align:center; font-size:120%; border-spacing: 0; padding: 0px;" cellpadding="10" | ||
| − | |[[# | + | |[[#Summary|Summary ]] |
| − | |[[# | + | |[[#What is 3D Slicer|What is 3D Slicer]] |
| − | |[[#Slicer Highlights|Slicer Highlights ]] | + | |[[#Slicer {{documentation/version}} Highlights|Slicer {{documentation/version}} Highlights]] |
|[[#Slicer Extensions|Slicer Extensions]] | |[[#Slicer Extensions|Slicer Extensions]] | ||
|[[#Other Improvements, Additions & Documentation| Other Improvements, Additions & Documentation]] | |[[#Other Improvements, Additions & Documentation| Other Improvements, Additions & Documentation]] | ||
| Line 37: | Line 37: | ||
The main [http://www.slicer.org slicer.org] pages provide a guided tour to the application, training materials, and the development community. New users should start there because we try to keep the pages organized and up to date. | The main [http://www.slicer.org slicer.org] pages provide a guided tour to the application, training materials, and the development community. New users should start there because we try to keep the pages organized and up to date. | ||
| + | <!-- | ||
Find out more about Slicer {{documentation/version}} in the webinar held on [...]: | Find out more about Slicer {{documentation/version}} in the webinar held on [...]: | ||
[[Image:Webinar-Slicer-4.1.png|link=http://vimeo.com/41096643|400px|center]] | [[Image:Webinar-Slicer-4.1.png|link=http://vimeo.com/41096643|400px|center]] | ||
| + | --> | ||
=What is 3D Slicer= | =What is 3D Slicer= | ||
| Line 57: | Line 59: | ||
Image:CLIAutoRun.png|Added AutoRun mode for CLI modules to interactively search in parameter space and to create pipelines of filters. Watch the [https://vimeo.com/64813350 demo]! | Image:CLIAutoRun.png|Added AutoRun mode for CLI modules to interactively search in parameter space and to create pipelines of filters. Watch the [https://vimeo.com/64813350 demo]! | ||
Image:Markups-GUI-Collapsed.png|The [[Documentation/{{documentation/version}}/Modules/Markups|Markups]] module replaces Annotation fiducials. | Image:Markups-GUI-Collapsed.png|The [[Documentation/{{documentation/version}}/Modules/Markups|Markups]] module replaces Annotation fiducials. | ||
| + | Image:RegLib_thumb.png| Slicer [[Documentation/{{documentation/version}}/Modules/BRAINSFit|image registration]] modules have been reimplemented using ITKv4, with streamlined interfaces and improved robustness. An updated [[Documentation/{{documentation/version}}/Registration/RegistrationLibrary| Registration Library]] is showcasing examples with downloadable data and full step-by-step instructions. | ||
</gallery> | </gallery> | ||
| Line 62: | Line 65: | ||
<gallery caption="New and Improved Extensions" widths="250px" heights="150px" perrow="4"> | <gallery caption="New and Improved Extensions" widths="250px" heights="150px" perrow="4"> | ||
| − | Image:AirwaySegmentation | + | Image:AirwaySegmentation-ICON.png|[[Documentation/{{documentation/version}}/Extensions/AirwaySegmentation|Airway Segmentation]] to segment the airway from chest CT images.{{new}} |
Image:CMRTK-logo.png|[[Documentation/{{documentation/version}}/Extensions/CARMA|Cardiac MRI Toolkit]] to analyze cardiac LGE-MRI images. {{new}} | Image:CMRTK-logo.png|[[Documentation/{{documentation/version}}/Extensions/CARMA|Cardiac MRI Toolkit]] to analyze cardiac LGE-MRI images. {{new}} | ||
| − | Image:ChangeTracker logo.png|[[Documentation/{{documentation/version}}/Extensions/ChangeTracker| Change Tracker]] for quantification of the subtle changes in pathology. {{new}} | + | Image:ChangeTracker logo.png|[[Documentation/{{documentation/version}}/Extensions/ChangeTracker| Change Tracker]] for quantification of the subtle changes in pathology.{{new}} |
| − | Image:DiceSimilarityCoefficient.png|[[Documentation/{{documentation/version}}/Extensions/DiceComputation|Dice Computation]] | + | Image:DataStoreIcon.png|[[Documentation/{{documentation/version}}/Extensions/DataStore|Data Store]] Allows to easily upload and download dataset files.{{new}} |
| − | Image:ErodeDilateLabelIcon.png|[[Documentation/{{documentation/version}}/Extensions/ErodeDilateLabel|Erode/Dilate Label]] to | + | Image:DiceSimilarityCoefficient.png|[[Documentation/{{documentation/version}}/Extensions/DiceComputation|Dice Computation]] to calculate the Dice Similarity Coefficient (DSC) between multiple label map images.{{new}} |
| − | Image:IASEM | + | Image:DTIAtlasBuilder.jpeg|[[Documentation/{{documentation/version}}/Extensions/DTIAtlasBuilder|DTI Atlas Builder]] to create an Atlas image as an average of several DTI images that will be registered.{{updated}} |
| + | Image:DTIPrep.png|[[Documentation/{{documentation/version}}/Extensions/DTIPrep|DTI Prep]].{{updated}} | ||
| + | Image:DTIProcess-mj.png|[[Documentation/{{documentation/version}}/Extensions/DTIProcess| DTI Process]].{{updated}} | ||
| + | Image:ErodeDilateLabelIcon.png|[[Documentation/{{documentation/version}}/Extensions/ErodeDilateLabel|Erode/Dilate Label]] to perform binary erosion and dilation on a specified label.{{new}} | ||
| + | Image:FacetedVisualizer.png|[[Documentation/{{documentation/version}}/Extensions/FacetedVisualizer|Faceted Visualizer]] to create flexible visualizations using Slicer by combining a 3D digital atlas and 3D scene models with an ontology.{{updated}} | ||
| + | Image:FiberViewerLight.png|[[Documentation/{{documentation/version}}/Extensions/FiberViewerLight| Fiber Viewer Light]] to create fiber clusters based on methods such as Length, Gravity, Hausdorff, or Mean Distribution and Normalized Cut.{{updated}} | ||
| + | Image:IASEM.png|[[Documentation/{{documentation/version}}/Extensions/IASEM| IASEM]] to segmentation and process of IASEM Electron Microscopy images.{{new}} | ||
Image:QuickToolsLogo.png|[[Documentation/{{documentation/version}}/Extensions/ImageMaker|Image Maker]] to create volumes from scratch. {{new}} | Image:QuickToolsLogo.png|[[Documentation/{{documentation/version}}/Extensions/ImageMaker|Image Maker]] to create volumes from scratch. {{new}} | ||
Image:LAScarSegmenter.png|[[Documentation/{{documentation/version}}/Extensions/LAScarSegmenter|LA Scar Segmenter]] to perform segmentation of the left atrial scarring tissue from MR images. {{new}} | Image:LAScarSegmenter.png|[[Documentation/{{documentation/version}}/Extensions/LAScarSegmenter|LA Scar Segmenter]] to perform segmentation of the left atrial scarring tissue from MR images. {{new}} | ||
| − | Image:LASegmenter.png|[[Documentation/{{documentation/version}}/Extensions/LASegmenter|LA Segmenter]] to perform segmentation of the left atrium from MR images. {{new}} | + | Image:LASegmenter.png|[[Documentation/{{documentation/version}}/Extensions/LASegmenter|LA Segmenter]] to perform segmentation of the left atrium from MR images.{{new}} |
| − | Image:LongitudinalPETCTLogo.png|[[Documentation/{{documentation/version}}/Extensions/LongitudinalPETCT| Longitudinal PET/CT]] to provide a user friendly Slicer interface for quantification of DICOM PET/CT image data {{updated} | + | Image:LongitudinalPETCTLogo.png|[[Documentation/{{documentation/version}}/Extensions/LongitudinalPETCT| Longitudinal PET/CT]] to provide a user friendly Slicer interface for quantification of DICOM PET/CT image data.{{updated} |
| − | Image:MatlabBridgeLogo.png|[[Documentation/{{documentation/version}}/Extensions/MatlabBridge| Matlab Bridge]] to allow running Matlab functions directly in 3D Slicer. {{new}} | + | Image:MatlabBridgeLogo.png|[[Documentation/{{documentation/version}}/Extensions/MatlabBridge| Matlab Bridge]] to allow running Matlab functions directly in 3D Slicer.{{new}} |
| − | Image: | + | Image:ModelClipIcon.png|[[Documentation/{{documentation/version}}/Extensions/ModelClip| Model Clip]] to set osteotomy trajectory with multiple planes, and clip the model with just one click.{{new}} |
| − | Image: | + | Image:PathPlannerIcon.png|[[Documentation/{{documentation/version}}/Extensions/PathPlanner| Path Planner]]to facilitate the creation of trajectory.{{new}} |
| − | Image:PlusRemoteLogo.png|[[Documentation/{{documentation/version}}/Extensions/PlusRemote| | + | Image:PkModeling.png|[[Documentation/{{documentation/version}}/Extensions/PkModeling| Pk Modeling]] to calculate quantitative parameters from Dynamic Contrast Enhanced DCE-MRI images.{{new}} |
| − | Image: | + | Image:PlusRemoteLogo.png|[[Documentation/{{documentation/version}}/Extensions/PlusRemote|Plus Remote]] to send commands through OpenIGTLink Remote to PLUS.{{new}} |
| − | Image:ReportingLogo.png|[[Documentation/{{documentation/version}}/Extensions/Reporting|Reporting]] to create image annotations/markup that are stored in a structured form. {{updated}} | + | Image:Portplacement_icon.png|[[Documentation/{{documentation/version}}/Extensions/PortPlacement| Port Placement]]to assists in the planning of surgical port placement in laparoscopic procedures.{{new}} |
| − | Image: | + | Image:ReportingLogo.png|[[Documentation/{{documentation/version}}/Extensions/Reporting|Reporting]] to create image annotations/markup that are stored in a structured form.{{updated}} |
| − | Image:SlicerIGTLogo.png|[[Documentation/{{documentation/version}}/Extensions/SlicerIGT|SlicerIGT]] to use all the advanced features of 3D Slicer for real-time navigation. {{new}} | + | Image:SkullStripper.png|[[Documentation/{{documentation/version}}/Extensions/SkullStripper|Skull Stripper]] to register an atlas image to the patient data.{{updated}} |
| − | Image:SlicerRT Logo 2.0 128x128.png|[[Documentation/{{documentation/version}}/Extensions/SlicerRT|SlicerRT]] a tool for powerful radiotherapy research. {{updated}} | + | Image:SlicerIGTLogo.png|[[Documentation/{{documentation/version}}/Extensions/SlicerIGT|SlicerIGT]] to use all the advanced features of 3D Slicer for real-time navigation.{{new}} |
| − | Image:SlicerToKiwiExporterLogo.png|[[Documentation/{{documentation/version}}/Extensions/SlicerToKiwiExporter| | + | Image:SlicerRT Logo 2.0 128x128.png|[[Documentation/{{documentation/version}}/Extensions/SlicerRT|SlicerRT]] is a tool for powerful radiotherapy research. {{updated}} |
| − | Image:SwissSkullStripper.png|[[Documentation/{{documentation/version}}/Extensions/SwissSkullStripper|Swiss Skull Stripper]] | + | Image:SlicerToKiwiExporterLogo.png|[[Documentation/{{documentation/version}}/Extensions/SlicerToKiwiExporter|Slicer To Kiwi Exporter]] to provide an easy way to export [[Documentation/{{documentation/version}}/Modules/Models|models]] into a [http://www.kiwiviewer.org/ KiwiViewer].{{new}} |
| − | Image:TrackerStabilizer.png|[[Documentation/{{documentation/version}}/Extensions/TrackerStabilizer|Tracker Stabilizer]] | + | Image:SobolevSegmenter.png|[[Documentation/{{documentation/version}}/Extensions/SobolevSegmenter|Sobolev Segmenter]] implements Sobolev inner product based active contour, using the Chan-Vese energy functional.{{updated}} |
| − | Image:TransformVisualizerLogo.png|[[Documentation/{{documentation/version}}/Extensions/TransformVisualizer|Transform Visualizer]] to visualize any transform (linear transform, B-spline deformable transform, any other non-linear transform) or vector volume. {{new}} | + | Image:SwissSkullStripper.png|[[Documentation/{{documentation/version}}/Extensions/SwissSkullStripper|Swiss Skull Stripper]] to register an atlas image to the patient data.{{new}} |
| − | Image:WindowLevelEffectLogo.png|[[Documentation/{{documentation/version}}/Extensions/WindowLevelEffect|Window/Level Effect]] to adjust window/level for volumes using mouse and/or region of interest. {{new}} | + | Image:TrackerStabilizer.png|[[Documentation/{{documentation/version}}/Extensions/TrackerStabilizer|Tracker Stabilizer]] to output a filtered transform node based on an tracker input (transform node).{{new}} |
| − | Image: | + | Image:TransformVisualizerLogo.png|[[Documentation/{{documentation/version}}/Extensions/TransformVisualizer|Transform Visualizer]] to visualize any transform (linear transform, B-spline deformable transform, any other non-linear transform) or vector volume.{{new}} |
| − | + | Image:TubeTK-scaled.png|[[Documentation/{{documentation/version}}/Extensions/TubeTK| TubeTK]] is an open-source toolkit for the segmentation, registration, and analysis of tubes and surfaces in images.{{updated}} | |
| − | + | Image:UKFTractography.png|[[Documentation/{{documentation/version}}/Extensions/UKFTractography|UKF Tractography]] a framework which uses an unscented Kalman filter for performing tractography.{{updated}} | |
| − | + | Image:VisuaLine.png|[[Documentation/{{documentation/version}}/Extensions/VisuaLine|VisuaLine]] to display lines in Slicer.{{updated}} | |
| + | Image:WindowLevelEffectLogo.png|[[Documentation/{{documentation/version}}/Extensions/WindowLevelEffect|Window/Level Effect]] to adjust window/level for volumes using mouse and/or region of interest.{{new}} | ||
| + | Image:XNATSlicerIcon.jpg|[[Documentation/{{documentation/version}}/Extensions/XNATSlicer| XNAT Slicer]] Secure GUI-based IO with any XNAT server.{{new}} | ||
| + | Image:PerkTutorLogo.png|[[Documentation/{{documentation/version}}/Extensions/PerkTutor|PerkTutor]] for training in image-guided needle interventions.{{new}} | ||
<!-- You could user either {{new}} or {{updated}} macros. | <!-- You could user either {{new}} or {{updated}} macros. | ||
| Line 99: | Line 111: | ||
Image:SlicerModels-Large.svg|Added advanced controls for the [[Documentation/{{documentation/version}}/Modules/Models|model display representation]] | Image:SlicerModels-Large.svg|Added advanced controls for the [[Documentation/{{documentation/version}}/Modules/Models|model display representation]] | ||
Image:SlicerModels-Large.svg|Depth peeling option for translucent surface models in [[Documentation/{{documentation/version}}/SlicerApplication/MainApplicationGUI#3D_Viewer|3D views]] | Image:SlicerModels-Large.svg|Depth peeling option for translucent surface models in [[Documentation/{{documentation/version}}/SlicerApplication/MainApplicationGUI#3D_Viewer|3D views]] | ||
| + | |||
| + | Image:Fiber select.PNG|Added [[Documentation/{{documentation/version}}/Modules/FiberBundleLabelSelect|fiber selection based on label map module]] | ||
| + | |||
| + | Image:TractographyDisplayModule.PNG|Added individual fiber editing option in [[Documentation/{{documentation/version}}/Modules/TractographyDisplay|Tractography Display module]] | ||
| + | |||
Image:SlicerVolume-Large.svg|Added read/write support for [[Documentation/{{documentation/version}}/SlicerApplication/SupportedDataFormat|MRC files]] | Image:SlicerVolume-Large.svg|Added read/write support for [[Documentation/{{documentation/version}}/SlicerApplication/SupportedDataFormat|MRC files]] | ||
Image:ctkDoubleSpinBox.gif|Improved support for image precision. | Image:ctkDoubleSpinBox.gif|Improved support for image precision. | ||
Latest revision as of 23:30, 4 October 2013
Home < Documentation < 4.3 < Announcements
|
For the latest Slicer documentation, visit the read-the-docs. |
| Summary | What is 3D Slicer | Slicer 4.3 Highlights | Slicer Extensions | Other Improvements, Additions & Documentation |
Summary
The community of Slicer developers is proud to announce the release of Slicer 4.3.
- Slicer 4.3 introduces
- a new App store, called the extension manager, for adding capabilities to Slicer. Close to 40 plug-ins are currently available.
- close to 500 feature improvements and bug fixes lead to improved performance and stability.
- augmentation of many modules.
- improved workflows in registration and dMRI
- the multivolume explorer allows interaction with time series and DCE data.
- major improvements to the DICOM module
- a new Markups module to streamline the use of fiducial lists
- Click here to download Slicer 4.3 for different platforms and find pointers to the source code, mailing lists and bug tracker.
- Please note that Slicer continues to be a research package and is not intended for clinical use. Testing of functionality is an ongoing activity with high priority, however, some features of Slicer are not fully tested.
- The Slicer Training page provides a series of courses for learning how to use Slicer. The portfolio contains self-guided presentation and sample data sets.
The main slicer.org pages provide a guided tour to the application, training materials, and the development community. New users should start there because we try to keep the pages organized and up to date.
What is 3D Slicer
Slicer is a community platform created for the purpose of subject specific image analysis and visualization.
- Multi-modality imaging including, MRI, CT, US, nuclear medicine, and microscopy
- Multi organ from head to toe
- Bidirectional interface for devices
- Expandable and interfaced to multiple toolkits
There is no restriction on use, but permissions and compliance with rules are responsibility of users. For details on the license see here
Slicer 4.3 Highlights
- New and Improved Toolkits and Modules
Added AutoRun mode for CLI modules to interactively search in parameter space and to create pipelines of filters. Watch the demo!
The Markups module replaces Annotation fiducials.
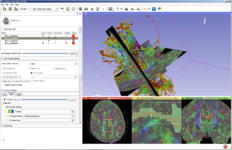
Slicer image registration modules have been reimplemented using ITKv4, with streamlined interfaces and improved robustness. An updated Registration Library is showcasing examples with downloadable data and full step-by-step instructions.
Slicer Extensions
- New and Improved Extensions
Airway Segmentation to segment the airway from chest CT images. NEW
Cardiac MRI Toolkit to analyze cardiac LGE-MRI images. NEW
Change Tracker for quantification of the subtle changes in pathology. NEW
Data Store Allows to easily upload and download dataset files. NEW
Dice Computation to calculate the Dice Similarity Coefficient (DSC) between multiple label map images. NEW
DTI Atlas Builder to create an Atlas image as an average of several DTI images that will be registered. UPDATED
DTI Prep. UPDATED
DTI Process. UPDATED
Erode/Dilate Label to perform binary erosion and dilation on a specified label. NEW
Faceted Visualizer to create flexible visualizations using Slicer by combining a 3D digital atlas and 3D scene models with an ontology. UPDATED
Fiber Viewer Light to create fiber clusters based on methods such as Length, Gravity, Hausdorff, or Mean Distribution and Normalized Cut. UPDATED
IASEM to segmentation and process of IASEM Electron Microscopy images. NEW
Image Maker to create volumes from scratch. NEW
LA Scar Segmenter to perform segmentation of the left atrial scarring tissue from MR images. NEW
LA Segmenter to perform segmentation of the left atrium from MR images. NEW
Longitudinal PET/CT to provide a user friendly Slicer interface for quantification of DICOM PET/CT image data.{{updated}
Matlab Bridge to allow running Matlab functions directly in 3D Slicer. NEW
Model Clip to set osteotomy trajectory with multiple planes, and clip the model with just one click. NEW
Path Plannerto facilitate the creation of trajectory. NEW
Pk Modeling to calculate quantitative parameters from Dynamic Contrast Enhanced DCE-MRI images. NEW
Plus Remote to send commands through OpenIGTLink Remote to PLUS. NEW
Port Placementto assists in the planning of surgical port placement in laparoscopic procedures. NEW
Reporting to create image annotations/markup that are stored in a structured form. UPDATED
Skull Stripper to register an atlas image to the patient data. UPDATED
SlicerIGT to use all the advanced features of 3D Slicer for real-time navigation. NEW
SlicerRT is a tool for powerful radiotherapy research. UPDATED
Slicer To Kiwi Exporter to provide an easy way to export models into a KiwiViewer. NEW
Sobolev Segmenter implements Sobolev inner product based active contour, using the Chan-Vese energy functional. UPDATED
Swiss Skull Stripper to register an atlas image to the patient data. NEW
Tracker Stabilizer to output a filtered transform node based on an tracker input (transform node). NEW
Transform Visualizer to visualize any transform (linear transform, B-spline deformable transform, any other non-linear transform) or vector volume. NEW
TubeTK is an open-source toolkit for the segmentation, registration, and analysis of tubes and surfaces in images. UPDATED
UKF Tractography a framework which uses an unscented Kalman filter for performing tractography. UPDATED
VisuaLine to display lines in Slicer. UPDATED
Window/Level Effect to adjust window/level for volumes using mouse and/or region of interest. NEW
XNAT Slicer Secure GUI-based IO with any XNAT server. NEW
PerkTutor for training in image-guided needle interventions. NEW
Other Improvements, Additions & Documentation
Added advanced controls for the model display representation
Depth peeling option for translucent surface models in 3D views
Added individual fiber editing option in Tractography Display module
Added read/write support for MRC files
For Developers: Looking at the Code Changes
From a git checkout you can easily see the all the commits since the time of the 4.4.2 release:
git log --since="Sat Dec 8 03:32:53 2012"
To see a summary of your own commits, you could use something like:
git log --since="Sat Dec 8 03:32:53 2012" --oneline --author=pieper
see the git log man page for more options.
Commit stats and full changelog