Difference between revisions of "DTI Fiber Clustering Module-Documentation-3.6.2"
From Slicer Wiki
(Created page with '===Module Name=== Fiber Clustering and Analysis (EM Fiber Clustering) Module {| |thumb|280px|Module interface |[[File:shot2.png|thumb|280px|Traject…') |
m |
||
| (10 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
{| | {| | ||
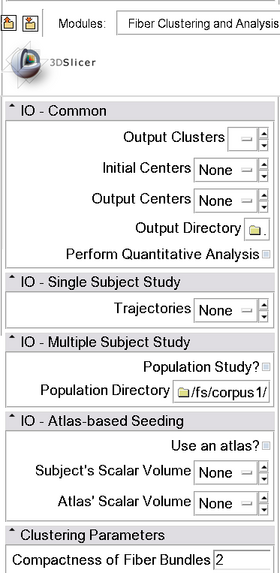
|[[File:moduleInterface.png|thumb|280px|Module interface]] | |[[File:moduleInterface.png|thumb|280px|Module interface]] | ||
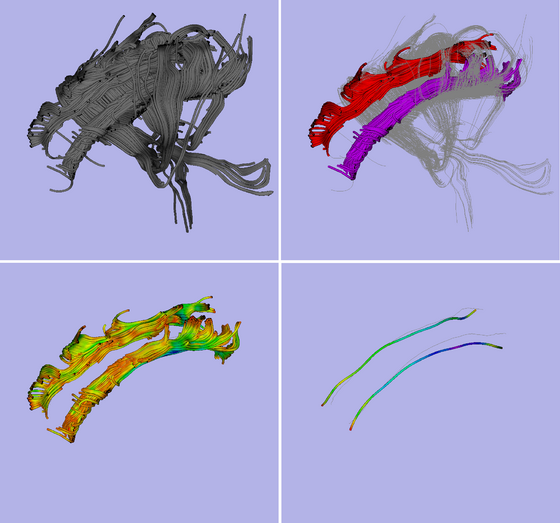
| − | |[[File: | + | |[[File:test1p.png|thumb|560px|An example of DTI fiber clustering and tract-oriented quantitative analysis]] |
|} | |} | ||
| + | |||
| + | ===Installation=== | ||
| + | |||
| + | To add the module to an installed binary of slicer (3.6.2 and later): | ||
| + | |||
| + | * Use the View->Extension Manager menu option | ||
| + | * The dialog will be initialized with the URL to the extensions that have been compiled to match your binary of slicer. | ||
| + | * Select '''EM Fiber Clustering''' to install and click to download. The installed extension, Fiber Clustering and Analysis, will be available when you restart slicer under Modules --> Diffusion --> Tractography. | ||
| + | |||
| + | Please report any installation problem to Mahnaz Maddah (mmaddah at alum.mit.edu) | ||
| + | |||
| + | ===Instructions and Test Data=== | ||
| + | |||
| + | == Single-Subject == | ||
| + | |||
| + | Clustering of the cingulum fiber tracts, Test1 Data: [[Media:single_subject.tar.gz]], Guide: [[Media:slide_test1.pdf]] | ||
| + | |||
| + | == Single-Subject with Atlas-Based Seeding == | ||
| + | |||
| + | Clustering of the uncinate fasciculus tract using the SRI24 atlas, Test2 Data: [[Media:atlas_based_seeding.tar.gz]], Guide: [[Media:slide_test2.pdf]] | ||
| + | |||
| + | == Multi-Subject == | ||
| + | |||
| + | Clustering of the genu in multiple subjects, Test3 Data: [[Media:multi-subject.tar.gz]], Guide: [[Media:slide_test3.pdf]] | ||
Latest revision as of 22:59, 31 January 2011
Home < DTI Fiber Clustering Module-Documentation-3.6.2Contents
Module Name
Fiber Clustering and Analysis (EM Fiber Clustering) Module
Installation
To add the module to an installed binary of slicer (3.6.2 and later):
- Use the View->Extension Manager menu option
- The dialog will be initialized with the URL to the extensions that have been compiled to match your binary of slicer.
- Select EM Fiber Clustering to install and click to download. The installed extension, Fiber Clustering and Analysis, will be available when you restart slicer under Modules --> Diffusion --> Tractography.
Please report any installation problem to Mahnaz Maddah (mmaddah at alum.mit.edu)
Instructions and Test Data
Single-Subject
Clustering of the cingulum fiber tracts, Test1 Data: Media:single_subject.tar.gz, Guide: Media:slide_test1.pdf
Single-Subject with Atlas-Based Seeding
Clustering of the uncinate fasciculus tract using the SRI24 atlas, Test2 Data: Media:atlas_based_seeding.tar.gz, Guide: Media:slide_test2.pdf
Multi-Subject
Clustering of the genu in multiple subjects, Test3 Data: Media:multi-subject.tar.gz, Guide: Media:slide_test3.pdf