Documentation/4.3/Extensions/SlicerRT
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
Authors: Csaba Pinter (PerkLab, Queen's University), Andras Lasso (PerkLab, Queen's University), Kevin Wang (Radiation Medicine Program, Princess Margaret Hospital, University Health Network Toronto)
Contributors: Greg Sharp (Massachusetts General Hospital), Steve Pieper (Isomics)
Contacts:
- Andras Lasso, <email>lasso@cs.queensu.ca</email>
- Mailing list, <email>slicerrt@alerts.assembla.com</email> (need to register to Assembla using the email address from which the email is sent)
- How to report an error
Website: slicerrt.org
License: Slicer license
Download/install: install 3D Slicer, start 3D Slicer, open the Extension Manager, install the SlicerRT extension (see more details on the download page)
Extension Description
|
Modules
Use Cases
|
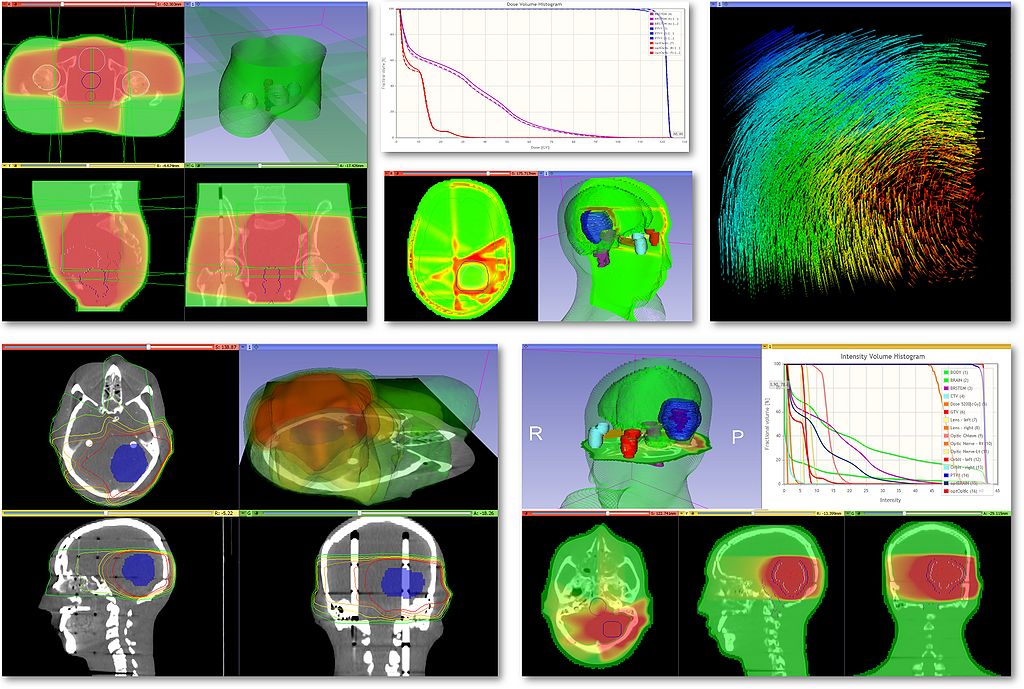
|
Tutorials
- Summer NA-MIC week 2013 tutorial (recommended)
- ECR 2013 - Medical University Vienna workshop
- Workshop material: download from NA-MIC.org
- RSNA 2012 tutorial
- Tutorial description: SlicerRT wiki: Slicer tutorials at RSNA 2012
- Sample data: download SlicerRT ART dose verification data from Midas server
- Summer NA-MIC week 2012 tutorial
Similar Extensions
- Plastimatch: SlicerRT and Plastimatch are complementary software libraries. Plastimatch focuses on delivering new computational methods radiotherapy, while SlicerRT aims for providing an easy-to-use interface for a wide range of stable, well-tested radiotherapy related features. SlicerRT uses Plastimatch internally for certain operations.
References
How to cite
Please cite the following paper when referring to Plus in your publication:
C. Pinter, A. Lasso, A. Wang, D. Jaffray and G. Fichtinger, "SlicerRT – Radiation therapy research toolkit for 3D Slicer", Med. Phys., 39(10) pp. 6332-6338, 2012
@ARTICLE{Pinter2012,
author = {Pinter, C. and Lasso, A. and Wang, A. and Jaffray, D. and Fichtinger, G.},
title = {SlicerRT – Radiation therapy research toolkit for 3D Slicer},
journal = {Med. Phys.},
year = {2012},
volume = {39},
number = {10},
pages = {6332-6338},
}



