Difference between revisions of "Modules:QDECModule-Documentation-3.4"
| Line 47: | Line 47: | ||
|- | |- | ||
| Set SUBJECTS_DIR | | Set SUBJECTS_DIR | ||
| − | | | + | | Set the SUBJECTS_DIR environment variable |
|- | |- | ||
| Load Table Data File | | Load Table Data File | ||
| Line 86: | Line 86: | ||
|} | |} | ||
* '''Display panel:''' | * '''Display panel:''' | ||
| + | |||
| + | |||
| + | {| border="1" | ||
| + | |- | ||
| + | | rowspan="2"| [[Image:SlicerQDECDisplay-3.4.jpg]] | ||
| + | |- | ||
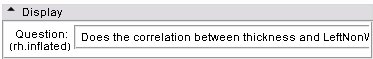
| + | | Question | ||
| + | | Pick which overlay is represented on the average brain, the overlays are generated in response to the question that was posed by the measures, for example, does the average thickness differ from zero? Selecting a new overlay also resets the colour node used to map the scalars on the model. You can manually select an overlay and color node in the Models Display panel if you wish. | ||
| + | |} | ||
== Development == | == Development == | ||
Revision as of 21:25, 25 February 2009
Home < Modules:QDECModule-Documentation-3.4Return to Slicer 3.4 Documentation
QDEC
QDEC
General Information
Module Type & Category
Type: Interactive
Category: Informatics
Authors, Collaborators & Contact
- Nicole Aucoin: Brigham and Women's Hospital
- Nick Schmansky: MGH
- Doug Greve: MGH
- Contact: Nicole Aucoin, nicole@bwh.harvard.edu
Module Description
The Qdec Module provides support for the QDEC program from MGH: Query, Design, Estimate, Contrast.
Usage
Examples, Use Cases & Tutorials
- This module is useful for visualising the output of FreeSurfer's GLM processing
- 2007 Project Week page
- Presentation from 2007
Quick Tour of Features and Use
Before you start: Set up a subjects directory and set your SUBJECTS_DIR environment variable (you'll need to restart Slicer3). In that directory, put the set of subjects you wish to analyse, along with the fsaverage subject. Then make a subdirectory qdec, and in it place your qdec.table.dat file along with any .level files describing the discrete factors (ie gender.levels will have 'male' and 'female' on two separate lines).
You can also load precomputed .qdec archives using the 'Load Results Data File' button. This requires unzip and rm, and you can point Slicer to these applications via the Application Settings interface (View->Application Settings).
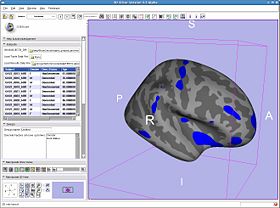
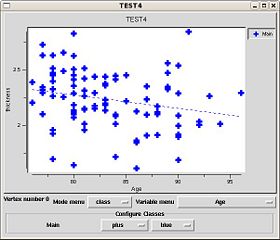
Once the results are loaded, you can plot results at various vertices by setting the setting the 3DViewer mouse mode to 'pick' using the toolbar button.
- Subjects panel:
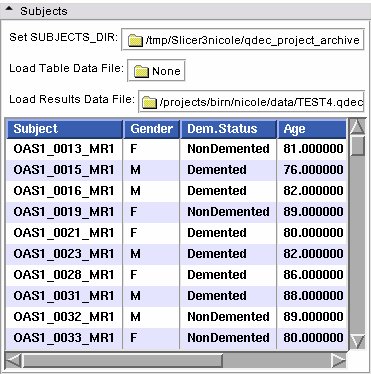
| |
| Set SUBJECTS_DIR | Set the SUBJECTS_DIR environment variable |
| Load Table Data File | Load a .dat file |
| Load Results Data File | Load a .qdec file |
| Table of Subjects | Filled in from the subjects on disk |
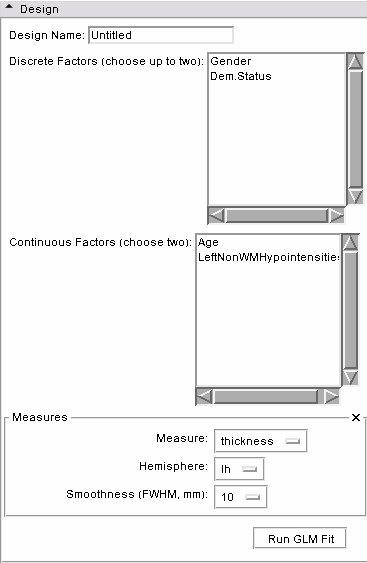
- Design panel:
- Display panel:
Development
Dependencies
The QDEC library (included with Slicer3) is required for this module. FreeSurfer is required for this module's use in calculating new GLM results, but is not necessary for loading pre computed ones.
Known bugs
Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Module Source Code:
- vtkQdecModuleGUI.cxx, vtkQdecModuleGUI.h
- vtkQdecModuleLogic.cxx, vtkQdecModuleLogic.h
- vtkGDFReader.cxx, vtkGDFReader.h
Library Source Code:
- QdecContrast.cpp, QdecContrast.h
- QdecDataTable.cpp, QdecDataTable.h
- QdecFactor.cpp, QdecFactor.h
- QdecGlmDesign.cpp, QdecGlmDesign.h
- QdecGlmFit.cpp, QdecGlmFit.h
- QdecGlmFitResults.cpp, QdecGlmFitResults.h
- QdecProject.cpp, QdecProject.h
- QdecSubject.cpp, QdecSubject.h
- QdecUtilities.cpp, QdecUtilities.h
- vtkFreeSurferReaders.tcl
Module Doxygen documentation:
Library Doxygen documentation:
More Information
Acknowledgment
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149.