Difference between revisions of "Documentation/4.8/Training"
Tag: 2017 source edit |
|||
| (14 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | <noinclude>{{documentation/ | + | <noinclude>{{documentation/historicaltraining}} |
| + | </noinclude> | ||
=Introduction: Slicer {{documentation/version}} Tutorials= | =Introduction: Slicer {{documentation/version}} Tutorials= | ||
| − | *This page contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment (version {{documentation/version}} release) to accomplish certain tasks. | + | *This page contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment (version {{documentation/version}} release) to accomplish certain tasks. |
*For tutorials for other versions of Slicer, please visit the [[Training| Slicer training portal]]. | *For tutorials for other versions of Slicer, please visit the [[Training| Slicer training portal]]. | ||
*For "reference manual" style documentation, please visit the [[Documentation/{{documentation/version}}|Slicer {{documentation/version}} documentation page]] | *For "reference manual" style documentation, please visit the [[Documentation/{{documentation/version}}|Slicer {{documentation/version}} documentation page]] | ||
| Line 8: | Line 9: | ||
| − | * Some of these tutorials are based on older releases of 3D Slicer, and are being upgraded to Slicer4.8. The concepts are still useful but bear in mind that some interface elements and features will be different in updated versions. | + | *Some of these tutorials are based on older releases of 3D Slicer, and are being upgraded to Slicer4.8. The concepts are still useful but bear in mind that some interface elements and features will be different in updated versions. |
__TOC__ | __TOC__ | ||
| − | =Quick Start Guide = | + | =Quick Start Guide= |
==Downloading and Installing Slicer== | ==Downloading and Installing Slicer== | ||
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
| − | *The [https://www.dropbox.com/s/ | + | *The [https://www.dropbox.com/s/gtir3x6e007snkx/QuickStartGuide_3DSlicer_SoniaPujol.pdf?dl=0| Quick Start Guide] shows how to install and start 3D Slicer |
*Author: Sonia Pujol, Ph.D. | *Author: Sonia Pujol, Ph.D. | ||
*Based on: 3D Slicer version 4.8 | *Based on: 3D Slicer version 4.8 | ||
| − | |align="right"| | + | | align="right" | |
[[image:QuickStart_image.png|250px|SlicerWelcome tutorial]] | [[image:QuickStart_image.png|250px|SlicerWelcome tutorial]] | ||
|} | |} | ||
| Line 27: | Line 28: | ||
==Slicer Welcome Tutorial== | ==Slicer Welcome Tutorial== | ||
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
*The [https://www.dropbox.com/s/vn8sqlof2kag2kk/SlicerWelcome-tutorial_Slicer4.8_SoniaPujol.pdf?dl=0| SlicerWelcome tutorial] is an introduction to Slicer based on the Welcome module. | *The [https://www.dropbox.com/s/vn8sqlof2kag2kk/SlicerWelcome-tutorial_Slicer4.8_SoniaPujol.pdf?dl=0| SlicerWelcome tutorial] is an introduction to Slicer based on the Welcome module. | ||
| Line 34: | Line 35: | ||
*Modules: Welcome to Slicer, Sample Data | *Modules: Welcome to Slicer, Sample Data | ||
*Based on: 3D Slicer version 4.8 | *Based on: 3D Slicer version 4.8 | ||
| − | |align="right"| | + | | align="right" | |
[[image:SlicerWelcome-image.png|250px|SlicerWelcome tutorial]] | [[image:SlicerWelcome-image.png|250px|SlicerWelcome tutorial]] | ||
|} | |} | ||
==Slicer4Minute Tutorial== | ==Slicer4Minute Tutorial== | ||
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
| − | *The [ | + | *The [https://www.dropbox.com/s/9jfsyhhgude5hf1/Slicer4.8minute_SoniaPujol.pdf?dl=0| Slicer4 Minute Tutorial] is a brief introduction to the advanced 3D visualization capabilities of Slicer 4.8. |
*Author: Sonia Pujol, Ph.D. | *Author: Sonia Pujol, Ph.D. | ||
| − | *Audience: First time users who want to discover Slicer in 4 minutes | + | *Audience: First-time users who want to discover Slicer in 4 minutes |
*Modules: Welcome to Slicer, Models | *Modules: Welcome to Slicer, Models | ||
| − | *Based on | + | *Based on 3D Slicer version 4.8 |
| − | *The [[ | + | *The [[Media:Slicer4minute.zip|Slicer4Minute dataset]] contains an MR scan of the brain and 3D reconstructions of the anatomy |
| − | |align="right"| | + | | align="right" | |
[[image:Slicer4minute-image.png|250px|right|Slicer4Minute tutorial]] | [[image:Slicer4minute-image.png|250px|right|Slicer4Minute tutorial]] | ||
|} | |} | ||
=3D Visualization= | =3D Visualization= | ||
| − | ==Slicer4 Data Loading and 3D Visualization == | + | ==Slicer4 Data Loading and 3D Visualization== |
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
| − | *The [[Media: | + | *The [[Media:3DDataLoadingandVisualization Slicer4.5 SoniaPujol.pdf| Data loading and 3D visualization]] course guides through the basics of loading and viewing volumes and 3D models in Slicer4 . |
*Author: Sonia Pujol, Ph.D. | *Author: Sonia Pujol, Ph.D. | ||
*Modules: Welcome to Slicer, Sample Data, Models. | *Modules: Welcome to Slicer, Sample Data, Models. | ||
| Line 61: | Line 62: | ||
*Based on: 3D Slicer version 4.5 | *Based on: 3D Slicer version 4.5 | ||
*The [http://slicer.kitware.com/midas3/download/?items=330421,1 3DVisualization dataset] contains an MR scan and a series of 3D models of the brain. | *The [http://slicer.kitware.com/midas3/download/?items=330421,1 3DVisualization dataset] contains an MR scan and a series of 3D models of the brain. | ||
| − | |align="right"| | + | | align="right" | |
[[Image:Slicer4DataLoading_tutorial.png|right|250px|]] | [[Image:Slicer4DataLoading_tutorial.png|right|250px|]] | ||
|} | |} | ||
==Slicer4 3D Visualization of DICOM images for Radiology Applications== | ==Slicer4 3D Visualization of DICOM images for Radiology Applications== | ||
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
| − | *The [https://www.dropbox.com/s/9jcjpl33qfh5pi6/3DVisualizationDICOM_Slicer4.8_SoniaPujol.pdf?dl=0 3D Visualization of DICOM images] course guides through 3D data loading and visualization of DICOM images for Radiology Applications in Slicer4. | + | *The [https://www.dropbox.com/s/9jcjpl33qfh5pi6/3DVisualizationDICOM_Slicer4.8_SoniaPujol.pdf?dl=0 3D Visualization of DICOM images] course guides through 3D data loading and visualization of DICOM images for Radiology Applications in Slicer4. |
*Author: Sonia Pujol, Ph.D., Kitt Shaffer, M.D., Ph.D., Ron Kikinis, M.D. | *Author: Sonia Pujol, Ph.D., Kitt Shaffer, M.D., Ph.D., Ron Kikinis, M.D. | ||
*Audience: Radiologists and users of Slicer who need a more comprehensive overview over Slicer4 visualization capabilities. | *Audience: Radiologists and users of Slicer who need a more comprehensive overview over Slicer4 visualization capabilities. | ||
*Modules: DICOM, Volumes, Volume Rendering, Models. | *Modules: DICOM, Volumes, Volume Rendering, Models. | ||
*Based on: 3D Slicer version 4.8 | *Based on: 3D Slicer version 4.8 | ||
| − | *The [[Media: | + | *The [[Media:3DVisualization DICOM images part1.zip| 3DVisualizationDICOM_part1]] and [[Media:3DVisualization DICOM images part2.zip| 3DVisualizationDICOM_part2]] datasets contain a series of MR and CT scans, and 3D models of the brain, lung and liver. |
| − | |align="right"| | + | | align="right" | |
[[Image:Slicer4RSNA_2.png|right|250px|]] | [[Image:Slicer4RSNA_2.png|right|250px|]] | ||
|} | |} | ||
| Line 80: | Line 81: | ||
=Programming= | =Programming= | ||
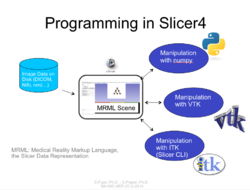
| − | == Slicer4 Programming Tutorial == | + | ==Slicer4 Programming Tutorial== |
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
| − | *The [https://www.dropbox.com/s/wrhrvvmplosiis1/Slicer4_ProgrammingTutorial_SPujol-SPieper_Nightly.pdf?dl=0# Slicer Programming tutorial] guides through the integration of a python module in Slicer4. | + | *The [https://www.dropbox.com/s/wrhrvvmplosiis1/Slicer4_ProgrammingTutorial_SPujol-SPieper_Nightly.pdf?dl=0# Slicer Programming tutorial] guides through the integration of a python module in Slicer4. |
*Author: Sonia Pujol, Ph.D., Steve Pieper, Ph.D. | *Author: Sonia Pujol, Ph.D., Steve Pieper, Ph.D. | ||
*Audience: Developers | *Audience: Developers | ||
*Based on: 3D Slicer version 4.7 | *Based on: 3D Slicer version 4.7 | ||
*The [https://www.dropbox.com/s/6yxu8qepmvywk0n/HelloPython_Nightly.zip?dl=0 HelloPython dataset] contains sample data set (MR scan of the brain) and complete Python module examples. | *The [https://www.dropbox.com/s/6yxu8qepmvywk0n/HelloPython_Nightly.zip?dl=0 HelloPython dataset] contains sample data set (MR scan of the brain) and complete Python module examples. | ||
| − | |align="right"| | + | | align="right" | |
[[Image:HelloPythonTutorial.png|right|250px|]] | [[Image:HelloPythonTutorial.png|right|250px|]] | ||
|} | |} | ||
| Line 95: | Line 96: | ||
==Developing and contributing extensions for 3D Slicer== | ==Developing and contributing extensions for 3D Slicer== | ||
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
| − | *The [http://goo.gl/IP4cdg Developing and contributing extensions for 3D Slicer tutorial ] is an introduction to the internals of 3D Slicer and the process of contributing a 3D Slicer extension. | + | *The [http://goo.gl/IP4cdg Developing and contributing extensions for 3D Slicer tutorial] is an introduction to the internals of 3D Slicer and the process of contributing a 3D Slicer extension. |
*Authors: Andrey Fedorov, Jean-Christophe Fillion-Robin, Steve Pieper | *Authors: Andrey Fedorov, Jean-Christophe Fillion-Robin, Steve Pieper | ||
*Audience: Developers | *Audience: Developers | ||
*Based on: 3D Slicer version 4.4 | *Based on: 3D Slicer version 4.4 | ||
| − | |align="right"| | + | | align="right" | |
[[Image:Contributing3DSlicerExtension.png|right|250px|]] | [[Image:Contributing3DSlicerExtension.png|right|250px|]] | ||
|} | |} | ||
=Segmentation= | =Segmentation= | ||
| − | == Slicer4 Image Segmentation == | + | ==Slicer4 Image Segmentation== |
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
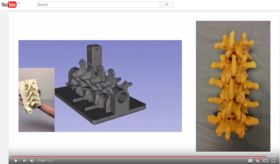
| − | * Segmentation for 3D printing: shows how to use the Segment Editor module for combining CAD designed parts with patient-specific models. | + | *Segmentation for 3D printing: shows how to use the Segment Editor module for combining CAD designed parts with patient-specific models. |
| − | ** '''[[Documentation/{{documentation/version}}/Training#Segmentation_for_3D_printing|Segmentation for 3D printing Step-by-step tutorial]]'''. Author: Csaba Pinter, MSc | + | **'''[[Documentation/{{documentation/version}}/Training#Segmentation_for_3D_printing|Segmentation for 3D printing Step-by-step tutorial]]'''. Author: Csaba Pinter, MSc |
| − | ** Audience: Users and developers interested in segmentation and 3D printing | + | **Audience: Users and developers interested in segmentation and 3D printing |
| − | ** Based on: 3D Slicer version 4.7 | + | **Based on: 3D Slicer version 4.7 |
| − | |align="right"|[[Image:20170717_3DPrintingTutorialYoutube.PNG|280px]] | + | | align="right" |[[Image:20170717_3DPrintingTutorialYoutube.PNG|280px]] |
|--- | |--- | ||
| | | | ||
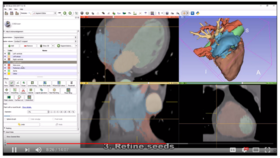
| − | * '''[https://youtu.be/BJoIexIvtGo Video tutorial: Whole heart segmentation from cardiac CT]''' shows how to use the Segment Editor module for segmenting heart ventricles, atria, and great vessels from cardiac CT volumes. | + | *'''[https://youtu.be/BJoIexIvtGo Video tutorial: Whole heart segmentation from cardiac CT]''' shows how to use the Segment Editor module for segmenting heart ventricles, atria, and great vessels from cardiac CT volumes. |
| − | ** Author: Andras Lasso, PhD | + | **Author: Andras Lasso, PhD |
| − | ** Audience: Users who need to segment heart structures, for example for visualization, quantification, or simulation. | + | **Audience: Users who need to segment heart structures, for example for visualization, quantification, or simulation. |
| − | ** [http://slicer.kitware.com/midas3/download/bitstream/738905/CTA-cardio2.nrrd Sample data set] | + | **[http://slicer.kitware.com/midas3/download/bitstream/738905/CTA-cardio2.nrrd Sample data set] |
| − | ** Based on: 3D Slicer version 4.8 | + | **Based on: 3D Slicer version 4.8 |
| − | |align="right"|[[Image:WholeHeartSegYoutube.png|280px]] | + | | align="right" |[[Image:WholeHeartSegYoutube.png|280px]] |
|} | |} | ||
=Registration= | =Registration= | ||
| − | == Slicer4 Image Registration == | + | ==Slicer4 Image Registration== |
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
*The [https://www.dropbox.com/s/chrugp2j2as5gop/ImageRegistration_Slicer4.8_SoniaPujol.pdf?dl=0 Registration tutorial] shows how to perform intra- and inter-subject registration within Slicer. | *The [https://www.dropbox.com/s/chrugp2j2as5gop/ImageRegistration_Slicer4.8_SoniaPujol.pdf?dl=0 Registration tutorial] shows how to perform intra- and inter-subject registration within Slicer. | ||
| − | * Authors: Sonia Pujol, Ph.D., Dominik Meier, Ph.D., Ron Kikinis, M.D. | + | *Authors: Sonia Pujol, Ph.D., Dominik Meier, Ph.D., Ron Kikinis, M.D. |
| − | * Audience: Users and developers interested in image registration | + | *Audience: Users and developers interested in image registration |
| − | * Dataset: [[Media:RegistrationData.zip| 3D Slicer Registration Data]] | + | *Dataset: [[Media:RegistrationData.zip| 3D Slicer Registration Data]] |
| − | |align="right"|[[File:registration_Slicer4.png|250px]] | + | | align="right" |[[File:registration_Slicer4.png|250px]] |
|} | |} | ||
| − | |||
| − | |||
| − | == Slicer Registration Case Library== | + | *Based on: 3D Slicer version 4.8; Compatible with Slicer4.10 |
| − | {|width="100%" | + | |
| + | ==Slicer Registration Case Library== | ||
| + | {| width="100%" | ||
| | | | ||
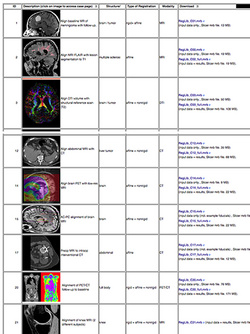
| − | *The ''[[Documentation/{{documentation/version}}/Registration/RegistrationLibrary|Slicer Registration Case Library]]'' provides | + | *The ''[[Documentation/{{documentation/version}}/Registration/RegistrationLibrary|Slicer Registration Case Library]]'' provides real-life example cases of using the Slicer registration tools. They include pre-computed dataset and step-by-step instructions for users to follow. |
| + | |||
:Author: Dominik Meier, Ph.D. | :Author: Dominik Meier, Ph.D. | ||
:Audience: users interested learning/applying Slicer image registration technology | :Audience: users interested learning/applying Slicer image registration technology | ||
| − | |align="right"|[[Image:RegLib_table.png|250px|link= | + | | align="right" |[[Image:RegLib_table.png|250px|link=https://www.slicer.org/wiki/Documentation/{{documentation/version}}/Registration/RegistrationLibrary]] |
| − | |} | + | |} =Slicer Extensions= |
| − | + | ==Slicer4 Diffusion Tensor Imaging Tutorial== | |
| − | =Slicer Extensions= | + | {| width="100%" |
| − | ==Slicer4 Diffusion Tensor Imaging Tutorial == | ||
| − | {|width="100%" | ||
| | | | ||
*Please visit [http://dmri.slicer.org/docs/ dmri.slicer.org/docs] for the latest documentation of SlicerDMRI. | *Please visit [http://dmri.slicer.org/docs/ dmri.slicer.org/docs] for the latest documentation of SlicerDMRI. | ||
| − | *The [https://www.dropbox.com/s/m7rzeblbdzr49bc/DiffusionMRIAnalysis_Slicer4.8_SoniaPujol.pdf?dl=0| Diffusion Tensor Imaging] course is an introduction to the basics of loading Diffusion Weighted images in Slicer, estimating tensors and generating fiber tracts. | + | *The [https://www.dropbox.com/s/m7rzeblbdzr49bc/DiffusionMRIAnalysis_Slicer4.8_SoniaPujol.pdf?dl=0| Diffusion Tensor Imaging] course is an introduction to the basics of loading Diffusion Weighted images in Slicer, estimating tensors and generating fiber tracts. |
*Author: Sonia Pujol, Ph.D. | *Author: Sonia Pujol, Ph.D. | ||
*Audience: End-users and developers | *Audience: End-users and developers | ||
*Modules: Data, Volumes, DWI to DTI Estimation, Diffusion Tensor Scalar Measurements, Editor, Markups,Tractography Label Map Seeding, Tractography Interactive Seeding | *Modules: Data, Volumes, DWI to DTI Estimation, Diffusion Tensor Scalar Measurements, Editor, Markups,Tractography Label Map Seeding, Tractography Interactive Seeding | ||
*Based on: 3D Slicer version 4.8 | *Based on: 3D Slicer version 4.8 | ||
| − | *The [[ | + | *The [[Media:Dti tutorial data.zip|DTI dataset]] contains an MR Diffusion Weighted Imaging scan of the brain. |
| − | |align="right"| | + | | align="right" | |
[[Image:Slicer4DTI Tutorial.png|right|250px|]] | [[Image:Slicer4DTI Tutorial.png|right|250px|]] | ||
|} | |} | ||
==Slicer4 Neurosurgical Planning Tutorial== | ==Slicer4 Neurosurgical Planning Tutorial== | ||
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
*Please visit [http://dmri.slicer.org/docs/ dmri.slicer.org/docs] for the latest documentation of SlicerDMRI. | *Please visit [http://dmri.slicer.org/docs/ dmri.slicer.org/docs] for the latest documentation of SlicerDMRI. | ||
| Line 174: | Line 174: | ||
*Based on: 3D Slicer version 4.6 | *Based on: 3D Slicer version 4.6 | ||
*The [[Media:WhiteMatterExplorationData.zip| White Matter Exploration dataset]] contains a Diffusion Weighted Imaging scan of brain tumor patient. | *The [[Media:WhiteMatterExplorationData.zip| White Matter Exploration dataset]] contains a Diffusion Weighted Imaging scan of brain tumor patient. | ||
| − | |align="right"| | + | | align="right" | |
[[Image:NeurosurgicalPlanningTutorial.png|right|250px|link=http://vimeo.com/67336069]] | [[Image:NeurosurgicalPlanningTutorial.png|right|250px|link=http://vimeo.com/67336069]] | ||
|} | |} | ||
| − | |||
==Slicer4 Quantitative Imaging tutorial== | ==Slicer4 Quantitative Imaging tutorial== | ||
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
| − | *The [[ | + | *The [[Media:QuantitativeImaging Slicer4.5.pdf| Slicer4 Quantitative Imaging tutorial]] guides through the use for Slicer for quantifying small volumetric changes in slow-growing tumors, and for calculating Standardized Uptake Value (SUV) from PET/CT data. |
*Authors: Sonia Pujol, Ph.D., Katarzyna Macura, M.D., Ron Kikinis, M.D. | *Authors: Sonia Pujol, Ph.D., Katarzyna Macura, M.D., Ron Kikinis, M.D. | ||
*Audience: Radiologists and users of Slicer who need a more comprehensive overview over Slicer4 quantitative imaging capabilities. | *Audience: Radiologists and users of Slicer who need a more comprehensive overview over Slicer4 quantitative imaging capabilities. | ||
*Modules: Data, Volumes, Models, Change Tracker, PET Standard Uptake Value Computation | *Modules: Data, Volumes, Models, Change Tracker, PET Standard Uptake Value Computation | ||
*Based on: 3D Slicer version 4.5 | *Based on: 3D Slicer version 4.5 | ||
| − | *The [[ | + | *The [[Media:QuantitativeImaging.zip| Quantitative Imaging dataset]] contains a series of MR and PET/CT data. |
| − | |align="right"| | + | | align="right" | |
[[Image:Slicer4_QuantitativeImaging.png|right|250px|]] | [[Image:Slicer4_QuantitativeImaging.png|right|250px|]] | ||
|} | |} | ||
| − | == Slicer4 IGT == | + | ==Slicer4 IGT== |
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
*[http://www.slicerigt.org/wp/user-tutorial/ Slicer IGT tutorials] | *[http://www.slicerigt.org/wp/user-tutorial/ Slicer IGT tutorials] | ||
| Line 201: | Line 200: | ||
*Modules: SlicerIGT Extension | *Modules: SlicerIGT Extension | ||
*Based on: Slicer4.3.1-2014.09.14 | *Based on: Slicer4.3.1-2014.09.14 | ||
| − | *Data: [https://onedrive.live.com/redir?resid=7230D4DEC6058018!2937&authkey=!AGQkSCZOwjVYXw8&ithint=folder%2cpptx | + | *Data: [https://onedrive.live.com/redir?resid=7230D4DEC6058018!2937&authkey=!AGQkSCZOwjVYXw8&ithint=folder%2cpptx Slicer-IGT datasets] |
| − | |align="right"| | + | | align="right" | |
[[Image:SlicetIGT.png|right|150px|]] | [[Image:SlicetIGT.png|right|150px|]] | ||
|} | |} | ||
| − | ==Slicer4 Radiation Therapy Tutorial == | + | ==Slicer4 Radiation Therapy Tutorial== |
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
| − | * The [https://github.com/SlicerRt/SlicerRtDoc/raw/master/tutorials/SlicerRT_WorldCongress_TutorialIGRT.pdf SlicerRT tutorial] is an introduction to the Radiation Therapy functionalities of Slicer. | + | *The [https://github.com/SlicerRt/SlicerRtDoc/raw/master/tutorials/SlicerRT_WorldCongress_TutorialIGRT.pdf SlicerRT tutorial] is an introduction to the Radiation Therapy functionalities of Slicer. |
| − | * Author: Csaba Pinter, Andras Lasso, An Wang, Gregory C. Sharp, David Jaffray, Gabor Fichtinger. | + | *Author: Csaba Pinter, Andras Lasso, An Wang, Gregory C. Sharp, David Jaffray, Gabor Fichtinger. |
| − | * Dataset: [http://slicer.kitware.com/midas3/download/item/205404/SlicerRT_WorldCongress_TutorialIGRT_Dataset.zip download] from MIDAS server | + | *Dataset: [http://slicer.kitware.com/midas3/download/item/205404/SlicerRT_WorldCongress_TutorialIGRT_Dataset.zip download] from MIDAS server |
| − | * Based on Slicer 4.7 | + | *Based on Slicer 4.7 |
| − | |align="right"| | + | | align="right" | |
<!-- [[Image:TUTORIAL-IMAGE-HERE.png|right|150px|]] --> | <!-- [[Image:TUTORIAL-IMAGE-HERE.png|right|150px|]] --> | ||
|} | |} | ||
| Line 221: | Line 220: | ||
==Slicer Pathology== | ==Slicer Pathology== | ||
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
*The [[Documentation/{{documentation/version}}/Extensions/SlicerPathology|Slicer Pathology Tutorial]] describes how to use the corresponding tools for automatic and semi-automatic pathology image segmentation. | *The [[Documentation/{{documentation/version}}/Extensions/SlicerPathology|Slicer Pathology Tutorial]] describes how to use the corresponding tools for automatic and semi-automatic pathology image segmentation. | ||
*Author: Erich Bremer (Stonybrook), Andriy Fedorov (Brigham and Women’s Hospital) | *Author: Erich Bremer (Stonybrook), Andriy Fedorov (Brigham and Women’s Hospital) | ||
*Dataset: Available directly with the Slicer Pathology Slicer extension. | *Dataset: Available directly with the Slicer Pathology Slicer extension. | ||
| − | |align="right"| | + | | align="right" | |
[[File:SlicerPathologyScreenShot8.png | 200px]]. | [[File:SlicerPathologyScreenShot8.png | 200px]]. | ||
|} | |} | ||
==SPHARM-PDM== | ==SPHARM-PDM== | ||
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
*The [https://www.nitrc.org/docman/view.php/308/1982/SPHARM-PDM_Tutorial_July2015.pdf SPHARM-PDM Tutorial] describes how to use SPHARM-PDM and ShapePopulationViewer Slicer extensions to respectively compute point-based models using a parametric boundary description for the computing of Shape Analysis and perform the quality control between the different models. | *The [https://www.nitrc.org/docman/view.php/308/1982/SPHARM-PDM_Tutorial_July2015.pdf SPHARM-PDM Tutorial] describes how to use SPHARM-PDM and ShapePopulationViewer Slicer extensions to respectively compute point-based models using a parametric boundary description for the computing of Shape Analysis and perform the quality control between the different models. | ||
*Author: Jonathan Perdomo (UNC), Beatriz Paniagua (Kitware Inc.) | *Author: Jonathan Perdomo (UNC), Beatriz Paniagua (Kitware Inc.) | ||
*Dataset: [https://www.nitrc.org/docman/view.php/308/1981/SPHARM_Tutorial_Data_July2015.zip Tutorial Data] | *Dataset: [https://www.nitrc.org/docman/view.php/308/1981/SPHARM_Tutorial_Data_July2015.zip Tutorial Data] | ||
| − | |align="right"| | + | | align="right" | |
[[File:SlicerWinterProjectWeek2017-SPHARM-PDM.png | 200px]]. | [[File:SlicerWinterProjectWeek2017-SPHARM-PDM.png | 200px]]. | ||
|} | |} | ||
==Fiber Bundle Volume Measurement== | ==Fiber Bundle Volume Measurement== | ||
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
*The [http://www.na-mic.org/Wiki/images/5/57/Fiber_Bundle_Volume_Measurement.pptx Fiber Bundle Volume Measurement Tutorial] aim is to calculate the volume of the fiber bundle that passes through the Corpus Callosum(CC). Following this tutorial, you’ll be able to (1) convert fiber bundles to label map and (2) calculate volume measurements from the fiber bundles. | *The [http://www.na-mic.org/Wiki/images/5/57/Fiber_Bundle_Volume_Measurement.pptx Fiber Bundle Volume Measurement Tutorial] aim is to calculate the volume of the fiber bundle that passes through the Corpus Callosum(CC). Following this tutorial, you’ll be able to (1) convert fiber bundles to label map and (2) calculate volume measurements from the fiber bundles. | ||
*Author: Shun Gong (Shanghai Changzheng Hospital, China) | *Author: Shun Gong (Shanghai Changzheng Hospital, China) | ||
*Dataset: [http://www.na-mic.org/Wiki/images/4/4c/FiberVolume_data.zip Tutorial data]: The following data are provided: Baseline image, Down sampled whole brain tractography (conducted as in the [[Documentation/{{documentation/version}}/Training#Slicer4_Diffusion_Tensor_Imaging_Tutorial|DWI tutorial]] and down-sampled to about 10000 fibers using Tractography Display module), Corpus callosum label map (drawn as in the [[Documentation/{{documentation/version}}/Training#Slicer4_Diffusion_Tensor_Imaging_Tutorial|DWI tutorial]]). | *Dataset: [http://www.na-mic.org/Wiki/images/4/4c/FiberVolume_data.zip Tutorial data]: The following data are provided: Baseline image, Down sampled whole brain tractography (conducted as in the [[Documentation/{{documentation/version}}/Training#Slicer4_Diffusion_Tensor_Imaging_Tutorial|DWI tutorial]] and down-sampled to about 10000 fibers using Tractography Display module), Corpus callosum label map (drawn as in the [[Documentation/{{documentation/version}}/Training#Slicer4_Diffusion_Tensor_Imaging_Tutorial|DWI tutorial]]). | ||
| − | |align="right"| | + | | align="right" | |
[[File:SlicerWinterProjectWeek2017-FiberBundleVolumeMeasurements.png | 200px]]. | [[File:SlicerWinterProjectWeek2017-FiberBundleVolumeMeasurements.png | 200px]]. | ||
|} | |} | ||
| − | = 3D Slicer version 4.7 Tutorial Contest= | + | =3D Slicer version 4.7 Tutorial Contest= |
For previous editions of the contest, please visit the [https://na-mic.org/wiki/Tutorial_Contests 3D Slicer Tutorial Contests page] | For previous editions of the contest, please visit the [https://na-mic.org/wiki/Tutorial_Contests 3D Slicer Tutorial Contests page] | ||
===Segmentation for 3D printing=== | ===Segmentation for 3D printing=== | ||
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
| − | *The [https://github.com/SlicerRt/SlicerRtDoc/raw/master/tutorials/SegmentationFor3DPrinting_TutorialContestWinter2017.pdf Segmentation for 3D printing Tutorial] ([https://github.com/SlicerRt/SlicerRtDoc/raw/master/tutorials/SegmentationFor3DPrinting_TutorialContestWinter2017.pptx pptx]) is an introduction to the new [[Documentation/{{documentation/version}}/Modules/SegmentEditor|Segment Editor]] module, demonstrated through the popular topic of 3D printing. | + | *The [https://github.com/SlicerRt/SlicerRtDoc/raw/master/tutorials/SegmentationFor3DPrinting_TutorialContestWinter2017.pdf Segmentation for 3D printing Tutorial] ([https://github.com/SlicerRt/SlicerRtDoc/raw/master/tutorials/SegmentationFor3DPrinting_TutorialContestWinter2017.pptx pptx]) is an introduction to the new [[Documentation/{{documentation/version}}/Modules/SegmentEditor|Segment Editor]] module, demonstrated through the popular topic of 3D printing. |
*Author: Csaba Pinter (Queen's University, Canada) | *Author: Csaba Pinter (Queen's University, Canada) | ||
| − | * [https://www.youtube.com/watch?v=Uht6Fwtr9hE Narrated video version on YouTube]. | + | *[https://www.youtube.com/watch?v=Uht6Fwtr9hE Narrated video version on YouTube]. |
*Dataset: [[:File:BasePiece.zip|Phantom base STL model]] Source: [http://perk-software.cs.queensu.ca/plus/doc/nightly/modelcatalog/ PerkLab]. | *Dataset: [[:File:BasePiece.zip|Phantom base STL model]] Source: [http://perk-software.cs.queensu.ca/plus/doc/nightly/modelcatalog/ PerkLab]. | ||
| − | |align="right"| | + | | align="right" | |
[[File:SlicerWinterProjectWeek2017-Segmentation-for-3d-printing.png | 200px]]. | [[File:SlicerWinterProjectWeek2017-Segmentation-for-3d-printing.png | 200px]]. | ||
|} | |} | ||
===Slicer Pathology=== | ===Slicer Pathology=== | ||
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
*The [[Documentation/{{documentation/version}}/Extensions/SlicerPathology|Slicer Pathology Tutorial]] describes how to use the corresponding tools for automatic and semi-automatic pathology image segmentation. | *The [[Documentation/{{documentation/version}}/Extensions/SlicerPathology|Slicer Pathology Tutorial]] describes how to use the corresponding tools for automatic and semi-automatic pathology image segmentation. | ||
*Author: Erich Bremer (Stonybrook), Andriy Fedorov (Brigham and Women’s Hospital) | *Author: Erich Bremer (Stonybrook), Andriy Fedorov (Brigham and Women’s Hospital) | ||
*Dataset: Available directly with the Slicer Pathology Slicer extension. | *Dataset: Available directly with the Slicer Pathology Slicer extension. | ||
| − | |align="right"| | + | | align="right" | |
[[File:SlicerPathologyScreenShot8.png | 200px]]. | [[File:SlicerPathologyScreenShot8.png | 200px]]. | ||
|} | |} | ||
===Simple Python Tool for Quality Control of DWI data=== | ===Simple Python Tool for Quality Control of DWI data=== | ||
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
*The [http://www.na-mic.org/Wiki/images/3/3a/SimpleDiffusionGradientInformationExtractorTutorial_Chauvin_Jan2017.pptx Simple Multi-shell Diffusion Gradients Information Extractor Tutorial] describes how to use a simple Python script for parsing multi-shell sensitizing gradients information from nifti file format (separated bvecs, bvals files). | *The [http://www.na-mic.org/Wiki/images/3/3a/SimpleDiffusionGradientInformationExtractorTutorial_Chauvin_Jan2017.pptx Simple Multi-shell Diffusion Gradients Information Extractor Tutorial] describes how to use a simple Python script for parsing multi-shell sensitizing gradients information from nifti file format (separated bvecs, bvals files). | ||
*Author: Laurent Chauvin (ETS Montreal) | *Author: Laurent Chauvin (ETS Montreal) | ||
*Dataset: Not available. | *Dataset: Not available. | ||
| − | |align="right"| | + | | align="right" | |
[[File:SlicerWinterProjectWeek2017-SimpleDiffusionGradientInformationExtractorTutorial.png | 200px]]. | [[File:SlicerWinterProjectWeek2017-SimpleDiffusionGradientInformationExtractorTutorial.png | 200px]]. | ||
|} | |} | ||
===SPHARM-PDM=== | ===SPHARM-PDM=== | ||
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
*The [https://www.nitrc.org/docman/view.php/308/1982/SPHARM-PDM_Tutorial_July2015.pdf SPHARM-PDM Tutorial] describes how to use SPHARM-PDM and ShapePopulationViewer Slicer extensions to respectively compute point-based models using a parametric boundary description for the computing of Shape Analysis and perform the quality control between the different models. | *The [https://www.nitrc.org/docman/view.php/308/1982/SPHARM-PDM_Tutorial_July2015.pdf SPHARM-PDM Tutorial] describes how to use SPHARM-PDM and ShapePopulationViewer Slicer extensions to respectively compute point-based models using a parametric boundary description for the computing of Shape Analysis and perform the quality control between the different models. | ||
*Author: Jonathan Perdomo (UNC), Beatriz Paniagua (Kitware Inc.) | *Author: Jonathan Perdomo (UNC), Beatriz Paniagua (Kitware Inc.) | ||
*Dataset: [https://www.nitrc.org/docman/view.php/308/1981/SPHARM_Tutorial_Data_July2015.zip Tutorial Data] | *Dataset: [https://www.nitrc.org/docman/view.php/308/1981/SPHARM_Tutorial_Data_July2015.zip Tutorial Data] | ||
| − | |align="right"| | + | | align="right" | |
[[File:SlicerWinterProjectWeek2017-SPHARM-PDM.png | 200px]]. | [[File:SlicerWinterProjectWeek2017-SPHARM-PDM.png | 200px]]. | ||
|} | |} | ||
===Integration of Robot Operating System (ROS) and 3D Slicer using OpenIGTLink=== | ===Integration of Robot Operating System (ROS) and 3D Slicer using OpenIGTLink=== | ||
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
*The [https://www.na-mic.org/Wiki/images/a/ab/ROSIGTLTutorial_Tokuda_Jan2017.pptx Integration of Robot Operating System (ROS) and 3D Slicer using OpenIGTLink Tutorial] describes the software architecture of surgical robot systems and allows to acquire hands-on experience of software-hardware integration for medical robotics. | *The [https://www.na-mic.org/Wiki/images/a/ab/ROSIGTLTutorial_Tokuda_Jan2017.pptx Integration of Robot Operating System (ROS) and 3D Slicer using OpenIGTLink Tutorial] describes the software architecture of surgical robot systems and allows to acquire hands-on experience of software-hardware integration for medical robotics. | ||
*Author: Junichi Tokuda (Brigham and Women’s Hospital) | *Author: Junichi Tokuda (Brigham and Women’s Hospital) | ||
*Dataset: Not available. | *Dataset: Not available. | ||
| − | |align="right"| | + | | align="right" | |
[[File:SlicerWinterProjectWeek2017-Integration-ROS-3DSlicer-OpenIGTLink.png | 200px]]. | [[File:SlicerWinterProjectWeek2017-Integration-ROS-3DSlicer-OpenIGTLink.png | 200px]]. | ||
|} | |} | ||
===Fiber Bundle Volume Measurement=== | ===Fiber Bundle Volume Measurement=== | ||
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
*The [http://www.na-mic.org/Wiki/images/5/57/Fiber_Bundle_Volume_Measurement.pptx Fiber Bundle Volume Measurement Tutorial] aim is to calculate the volume of the fiber bundle that passes through the Corpus Callosum(CC). Following this tutorial, you’ll be able to (1) convert fiber bundles to label map and (2) calculate volume measurements from the fiber bundles. | *The [http://www.na-mic.org/Wiki/images/5/57/Fiber_Bundle_Volume_Measurement.pptx Fiber Bundle Volume Measurement Tutorial] aim is to calculate the volume of the fiber bundle that passes through the Corpus Callosum(CC). Following this tutorial, you’ll be able to (1) convert fiber bundles to label map and (2) calculate volume measurements from the fiber bundles. | ||
*Author: Shun Gong (Shanghai Changzheng Hospital, China) | *Author: Shun Gong (Shanghai Changzheng Hospital, China) | ||
*Dataset: [http://www.na-mic.org/Wiki/images/4/4c/FiberVolume_data.zip Tutorial data]: The following data are provided: Baseline image, Down sampled whole brain tractography (conducted as in the [[Documentation/{{documentation/version}}/Training#Slicer4_Diffusion_Tensor_Imaging_Tutorial|DWI tutorial]] and down-sampled to about 10000 fibers using Tractography Display module), Corpus callosum label map (drawn as in the [[Documentation/{{documentation/version}}/Training#Slicer4_Diffusion_Tensor_Imaging_Tutorial|DWI tutorial]]). | *Dataset: [http://www.na-mic.org/Wiki/images/4/4c/FiberVolume_data.zip Tutorial data]: The following data are provided: Baseline image, Down sampled whole brain tractography (conducted as in the [[Documentation/{{documentation/version}}/Training#Slicer4_Diffusion_Tensor_Imaging_Tutorial|DWI tutorial]] and down-sampled to about 10000 fibers using Tractography Display module), Corpus callosum label map (drawn as in the [[Documentation/{{documentation/version}}/Training#Slicer4_Diffusion_Tensor_Imaging_Tutorial|DWI tutorial]]). | ||
| − | |align="right"| | + | | align="right" | |
[[File:SlicerWinterProjectWeek2017-FiberBundleVolumeMeasurements.png | 200px]]. | [[File:SlicerWinterProjectWeek2017-FiberBundleVolumeMeasurements.png | 200px]]. | ||
|} | |} | ||
| Line 317: | Line 316: | ||
=YouTube videos= | =YouTube videos= | ||
| − | {|width=" | + | |
| − | | | + | {| border="1" cellpadding="5" width="1200px" |
| − | + | | style="width:33%" |[[Image:Verterbra.png|right|250px|]] [https://www.youtube.com/watch?v=Uht6Fwtr9hE How to segment multiple vertebrae in spine CT for 3D printing - Author: Hillary Lia] | |
| − | + | | style="width:33%" |[[Image:Femurmodel.png|right|250px|]] [https://www.youtube.com/watch?v=0at15gjk-Ns Creating a femur model from CT volume using 3D Slicer - Author: Nabgha Farhat] | |
| − | + | | style="width:33%" |[[Image:3DPrinting.png|right|250px|]] [https://www.youtube.com/watch?v=MKLWzD0PiIc Preparing data for 3D printing - Author: PerkLab] | |
| − | | | + | |- |
| − | [[Image: | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | | | ||
| − | [[Image:3DPrinting.png|right|250px|]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | | | ||
| − | |||
|} | |} | ||
| − | Additional [http://www.youtube.com/results?search_query=3d+slicer&sm=3 | + | Additional [http://www.youtube.com/results?search_query=3d+slicer&sm=3 non-curated videos-based demonstrations using 3D Slicer are accessible on YouTube]. |
| − | =Teams Contributions = | + | =Teams Contributions= |
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
| − | * This ''Slicer 4.1 [http://vimeo.com/41096643 webinar]'' presents the new features and improvements of the release, and a brief overview of work for the next release. | + | *This ''Slicer 4.1 [http://vimeo.com/41096643 webinar]'' presents the new features and improvements of the release, and a brief overview of work for the next release. |
| − | * Authors: Steve Pieper Ph.D. | + | *Authors: Steve Pieper Ph.D. |
| − | * Audience: First time users and developers interested in Slicer 4.1 new features. | + | *Audience: First time users and developers interested in Slicer 4.1 new features. |
| − | * Length: 0h20m | + | *Length: 0h20m |
| − | |align="right"|[[Image:Webinar-Slicer-4.1.png|250px]] | + | | align="right" |[[Image:Webinar-Slicer-4.1.png|250px]] |
|} | |} | ||
<br> | <br> | ||
---- | ---- | ||
<br> | <br> | ||
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
*This ''Intro to Slicer 4.0 [http://vimeo.com/37671358 webinar]'' provides an introduction to 3DSlicer, and demonstrates core functionalities such as loading, visualizing and saving data. Basic processing tools, including manual registration, manual segmentation and tractography tools are also highlighted. This webinar is a general overview. For in depth information see the modules above and the documentation pages. | *This ''Intro to Slicer 4.0 [http://vimeo.com/37671358 webinar]'' provides an introduction to 3DSlicer, and demonstrates core functionalities such as loading, visualizing and saving data. Basic processing tools, including manual registration, manual segmentation and tractography tools are also highlighted. This webinar is a general overview. For in depth information see the modules above and the documentation pages. | ||
| − | *Authors: Julien Finet, M.S., Steve Pieper, Ph.D., Jean-Christophe Fillion-Robin, M.S. | + | *Authors: Julien Finet, M.S., Steve Pieper, Ph.D., Jean-Christophe Fillion-Robin, M.S. |
*Audience: First time users interested in a broad overview of Slicer’s features and tools. | *Audience: First time users interested in a broad overview of Slicer’s features and tools. | ||
*Length: 1h20m | *Length: 1h20m | ||
| − | |align="right"|[[Image:Webinar.png|250px]] | + | | align="right" |[[Image:Webinar.png|250px]] |
|} | |} | ||
<br> | <br> | ||
| Line 373: | Line 351: | ||
See the collection of videos on the [http://vimeo.com/album/2363361 Kitware vimeo album]. | See the collection of videos on the [http://vimeo.com/album/2363361 Kitware vimeo album]. | ||
| − | = External Resources = | + | =External Resources= |
| − | == International resources== | + | ==International resources== |
| − | ==Resources in Chinese == | + | ==Resources in Chinese== |
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
A 3D Slicer community on WeChat in China offers many tutorials and clinical examples in Chinese. Note that the images are of interest to non-Chinese speakers and Google Translate does a reasonable job of translating some of the text. | A 3D Slicer community on WeChat in China offers many tutorials and clinical examples in Chinese. Note that the images are of interest to non-Chinese speakers and Google Translate does a reasonable job of translating some of the text. | ||
| − | * [https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247486116&idx=1&sn=772e9d431ac32cbb73d08cf0e6bc219a&chksm=eacc0096ddbb89805d93ac4be181d1a35058031bac673d7a91b3b44dccee2bfd1d8461397635#rd Getting started 大脑前动脉远端动脉瘤手术夹闭治疗] | + | *[https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247486116&idx=1&sn=772e9d431ac32cbb73d08cf0e6bc219a&chksm=eacc0096ddbb89805d93ac4be181d1a35058031bac673d7a91b3b44dccee2bfd1d8461397635#rd Getting started 大脑前动脉远端动脉瘤手术夹闭治疗] |
| − | * [https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247484787&idx=1&sn=1f6279bdccab168fc79b7275e9fe91ca&chksm=eacc0f41ddbb8657be92f617661133d87bb55a4ecf12f786e97a8b7d5249a05d11e0cd620c3f#rd distal anterior cerebral artery aneurysm 3D Slicer:漂亮得不像实力派] | + | *[https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247484787&idx=1&sn=1f6279bdccab168fc79b7275e9fe91ca&chksm=eacc0f41ddbb8657be92f617661133d87bb55a4ecf12f786e97a8b7d5249a05d11e0cd620c3f#rd distal anterior cerebral artery aneurysm 3D Slicer:漂亮得不像实力派] |
| − | * [https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247486360&idx=1&sn=f833b13a26f543aa9175419a03df7f52&chksm=eacc01aaddbb88bcb004773a4db8a9b3c7633d21cda3956f84b96515252eb861c5eb1e75a60b&mpshare=1&scene=24&srcid=0212LIfOkBRm9CvA7ImHCpRt#rd meningioma skull resection 脑膜瘤患者颅骨切除一期修补的3DSlicer方案] | + | *[https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247486360&idx=1&sn=f833b13a26f543aa9175419a03df7f52&chksm=eacc01aaddbb88bcb004773a4db8a9b3c7633d21cda3956f84b96515252eb861c5eb1e75a60b&mpshare=1&scene=24&srcid=0212LIfOkBRm9CvA7ImHCpRt#rd meningioma skull resection 脑膜瘤患者颅骨切除一期修补的3DSlicer方案] |
| − | * [https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247486079&idx=1&sn=9b926dc398a408e3441082b9e0ffde61&chksm=eacc004dddbb895bf9b60f5f1bc443513196e4cb90a6caf6f348a4da7b7fc22eb658661aeb49&mpshare=1&scene=24&srcid=0212AFtT2Wq7K7bvkMGTdyih#rd Cerebral hemorrhage by forehead positioning method 脑出血经额手术定位法(五 ] | + | *[https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247486079&idx=1&sn=9b926dc398a408e3441082b9e0ffde61&chksm=eacc004dddbb895bf9b60f5f1bc443513196e4cb90a6caf6f348a4da7b7fc22eb658661aeb49&mpshare=1&scene=24&srcid=0212AFtT2Wq7K7bvkMGTdyih#rd Cerebral hemorrhage by forehead positioning method 脑出血经额手术定位法(五] |
| − | * [https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247485356&idx=1&sn=044f5899b651b35994db00c32ab688ee&chksm=eacc0d9eddbb8488f16ff82bb1dda8456a4011790fed024781972d578783e67781443cf4a319&mpshare=1&scene=24&srcid=0212G45TadrPnX8tp9eaNXUs#rd Hematoma modeling 血肿建模的第11种方法] | + | *[https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247485356&idx=1&sn=044f5899b651b35994db00c32ab688ee&chksm=eacc0d9eddbb8488f16ff82bb1dda8456a4011790fed024781972d578783e67781443cf4a319&mpshare=1&scene=24&srcid=0212G45TadrPnX8tp9eaNXUs#rd Hematoma modeling 血肿建模的第11种方法] |
| − | * [https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247486025&idx=1&sn=b281324893be4ab116d20826f1b426c3&chksm=eacc007bddbb896d9deb096f209278f40c0b52c6410a8a9ff3ce8c3697c99304f18eb678f11e&mpshare=1&scene=24&srcid=02125v1kxvIGmfkxx7mUZcCM#rd Mobile phone positioning and AR application 手机定位及AR应用的初步探索] | + | *[https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247486025&idx=1&sn=b281324893be4ab116d20826f1b426c3&chksm=eacc007bddbb896d9deb096f209278f40c0b52c6410a8a9ff3ce8c3697c99304f18eb678f11e&mpshare=1&scene=24&srcid=02125v1kxvIGmfkxx7mUZcCM#rd Mobile phone positioning and AR application 手机定位及AR应用的初步探索] |
| − | |align="right"| | + | | align="right" | |
[[image:Wechat-hemorage-2018-02-12.png|250px|Example WeChat tutorial slides]] | [[image:Wechat-hemorage-2018-02-12.png|250px|Example WeChat tutorial slides]] | ||
|} | |} | ||
| − | ==Resources in German == | + | ==Resources in German== |
| − | * [https://www.youtube.com/watch?v=sl-00kGpuPk&list=PLJWCUXz3GeAfmYLiFcKus_c0jcsMnVsgb A series of four YouTube videos on python programming in Slicer] (German narration with English subtitles) | + | *[https://www.youtube.com/watch?v=sl-00kGpuPk&list=PLJWCUXz3GeAfmYLiFcKus_c0jcsMnVsgb A series of four YouTube videos on python programming in Slicer] (German narration with English subtitles) |
| − | == Murat Maga's blog posts about using 3D Slicer for biology == | + | ==Murat Maga's blog posts about using 3D Slicer for biology== |
| − | * [https://blogs.uw.edu/maga/2017/04/11/getting-started-with-3d-slicer-as-a-biologist/ Slicer for Biologists] | + | *[https://blogs.uw.edu/maga/2017/04/11/getting-started-with-3d-slicer-as-a-biologist/ Slicer for Biologists] |
| − | * [https://blogs.uw.edu/maga/2017/04/11/a-worked-example-getting-and-visualizing-data-from-digimorph/ Loading data from DigiMorph] | + | *[https://blogs.uw.edu/maga/2017/04/11/a-worked-example-getting-and-visualizing-data-from-digimorph/ Loading data from DigiMorph] |
| − | * [https://blogs.uw.edu/maga/2017/04/11/morphosource-data-and-dealing-with-dicom-series-in-slicer/ Fixing problem DICOM] | + | *[https://blogs.uw.edu/maga/2017/04/11/morphosource-data-and-dealing-with-dicom-series-in-slicer/ Fixing problem DICOM] |
| − | * [https://blogs.uw.edu/maga/2017/04/12/scissors-tool-is-awesome/ Scissors tool is awesom] | + | *[https://blogs.uw.edu/maga/2017/04/12/scissors-tool-is-awesome/ Scissors tool is awesom] |
| − | == Using the (legacy) Editor == | + | ==Using the (legacy) Editor== |
| − | === Fast GrowCut === | + | ===Fast GrowCut=== |
| − | {|width="100%" | + | {| width="100%" |
| | | | ||
| − | * The [[ | + | *The [[Media:FastGrowCutTutorial.pdf|Fast GrowCut tutorial]] shows how to perform a segmentation using the Fast GrowCut effect in Slicer. |
| − | * Authors: Hillary Lia | + | *Authors: Hillary Lia |
| − | * Audience: Users interested in segmentation | + | *Audience: Users interested in segmentation |
| − | |align="right"|[[File:FastGrowCutLogo.png|200px]] | + | | align="right" |[[File:FastGrowCutLogo.png|200px]] |
|} | |} | ||
| − | === Use case: Slicer in paleontology === | + | ===Use case: Slicer in paleontology=== |
This set of tutorials about the use of slicer in paleontology is very well written and provides step-by-step instructions. Even though it covers slicer version 3.4, many of the concepts and techniques have applicability to the new version and to any 3D imaging field: | This set of tutorials about the use of slicer in paleontology is very well written and provides step-by-step instructions. Even though it covers slicer version 3.4, many of the concepts and techniques have applicability to the new version and to any 3D imaging field: | ||
| − | * [http://openpaleo.blogspot.com/2008/12/3d-slicer-tutorial.html Open Source Paleontologist: 3D Slicer: The Tutorial] | + | *[http://openpaleo.blogspot.com/2008/12/3d-slicer-tutorial.html Open Source Paleontologist: 3D Slicer: The Tutorial] |
| − | * [http://openpaleo.blogspot.com/2008/12/3d-slicer-tutorial-part-ii.html Open Source Paleontologist: 3D Slicer: The Tutorial Part II] | + | *[http://openpaleo.blogspot.com/2008/12/3d-slicer-tutorial-part-ii.html Open Source Paleontologist: 3D Slicer: The Tutorial Part II] |
| − | * [http://openpaleo.blogspot.com/2008/12/3d-slicer-tutorial-part-iii.html Open Source Paleontologist: 3D Slicer: The Tutorial Part III] | + | *[http://openpaleo.blogspot.com/2008/12/3d-slicer-tutorial-part-iii.html Open Source Paleontologist: 3D Slicer: The Tutorial Part III] |
| − | * [http://openpaleo.blogspot.com/2008/12/3d-slicer-tutorial-part-iv.html Open Source Paleontologist: 3D Slicer: The Tutorial Part IV] | + | *[http://openpaleo.blogspot.com/2008/12/3d-slicer-tutorial-part-iv.html Open Source Paleontologist: 3D Slicer: The Tutorial Part IV] |
| − | * [http://openpaleo.blogspot.com/2009/03/3d-slicer-tutorial-part-v.html Open Source Paleontologist: 3D Slicer: The Tutorial Part V] | + | *[http://openpaleo.blogspot.com/2009/03/3d-slicer-tutorial-part-v.html Open Source Paleontologist: 3D Slicer: The Tutorial Part V] |
| − | * [http://openpaleo.blogspot.com/2009/03/3d-slicer-tutorial-part-vi.html Open Source Paleontologist: 3D Slicer: The Tutorial Part VI] | + | *[http://openpaleo.blogspot.com/2009/03/3d-slicer-tutorial-part-vi.html Open Source Paleontologist: 3D Slicer: The Tutorial Part VI] |
Latest revision as of 22:12, 22 November 2022
Home < Documentation < 4.8 < Training
 |
This section is currently out-of-date and may contain errors but is retained for historical reference. Up-to-date training materials can be found at Documentation/Nightly/Training |
Introduction: Slicer 4.8 Tutorials
- This page contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment (version 4.8 release) to accomplish certain tasks.
- For tutorials for other versions of Slicer, please visit the Slicer training portal.
- For "reference manual" style documentation, please visit the Slicer 4.8 documentation page
- For questions related to the Slicer4 Training Compendium, please send an e-mail to Sonia Pujol, Ph.D., Director of Training of 3D Slicer.
- Some of these tutorials are based on older releases of 3D Slicer, and are being upgraded to Slicer4.8. The concepts are still useful but bear in mind that some interface elements and features will be different in updated versions.
Contents
- 1 Introduction: Slicer 4.8 Tutorials
- 2 Quick Start Guide
- 3 General Introduction
- 4 3D Visualization
- 5 Programming
- 6 Segmentation
- 7 Registration
- 7.1 Slicer4 Image Registration
- 7.2 Slicer Registration Case Library
- 7.3 Slicer4 Diffusion Tensor Imaging Tutorial
- 7.4 Slicer4 Neurosurgical Planning Tutorial
- 7.5 Slicer4 Quantitative Imaging tutorial
- 7.6 Slicer4 IGT
- 7.7 Slicer4 Radiation Therapy Tutorial
- 7.8 Slicer Pathology
- 7.9 SPHARM-PDM
- 7.10 Fiber Bundle Volume Measurement
- 8 3D Slicer version 4.7 Tutorial Contest
- 9 YouTube videos
- 10 Teams Contributions
- 11 External Resources
Quick Start Guide
Downloading and Installing Slicer
|
General Introduction
Slicer Welcome Tutorial
|
Slicer4Minute Tutorial
|
3D Visualization
Slicer4 Data Loading and 3D Visualization
|
Slicer4 3D Visualization of DICOM images for Radiology Applications
|
Programming
Slicer4 Programming Tutorial
|
For additional Python scripts examples, please visit the Script Repository page
Developing and contributing extensions for 3D Slicer
|
Segmentation
Slicer4 Image Segmentation
|
|
|
|
Registration
Slicer4 Image Registration
|

|
- Based on: 3D Slicer version 4.8; Compatible with Slicer4.10
Slicer Registration Case Library
|
|
=Slicer Extensions=
Slicer4 Diffusion Tensor Imaging Tutorial
|
Slicer4 Neurosurgical Planning Tutorial
|
Slicer4 Quantitative Imaging tutorial
|
Slicer4 IGT
|
Slicer4 Radiation Therapy Tutorial
|
Slicer Pathology
|
SPHARM-PDM
|
Fiber Bundle Volume Measurement
|
3D Slicer version 4.7 Tutorial Contest
For previous editions of the contest, please visit the 3D Slicer Tutorial Contests page
Segmentation for 3D printing
|
Slicer Pathology
|
Simple Python Tool for Quality Control of DWI data
|
SPHARM-PDM
|
Integration of Robot Operating System (ROS) and 3D Slicer using OpenIGTLink
|
Fiber Bundle Volume Measurement
|
YouTube videos
Additional non-curated videos-based demonstrations using 3D Slicer are accessible on YouTube.
Teams Contributions
|
|
|
|
See the collection of videos on the Kitware vimeo album.
External Resources
International resources
Resources in Chinese
|
A 3D Slicer community on WeChat in China offers many tutorials and clinical examples in Chinese. Note that the images are of interest to non-Chinese speakers and Google Translate does a reasonable job of translating some of the text. |
Resources in German
- A series of four YouTube videos on python programming in Slicer (German narration with English subtitles)
Murat Maga's blog posts about using 3D Slicer for biology
Using the (legacy) Editor
Fast GrowCut
|

|
Use case: Slicer in paleontology
This set of tutorials about the use of slicer in paleontology is very well written and provides step-by-step instructions. Even though it covers slicer version 3.4, many of the concepts and techniques have applicability to the new version and to any 3D imaging field:
- Open Source Paleontologist: 3D Slicer: The Tutorial
- Open Source Paleontologist: 3D Slicer: The Tutorial Part II
- Open Source Paleontologist: 3D Slicer: The Tutorial Part III
- Open Source Paleontologist: 3D Slicer: The Tutorial Part IV
- Open Source Paleontologist: 3D Slicer: The Tutorial Part V
- Open Source Paleontologist: 3D Slicer: The Tutorial Part VI