Difference between revisions of "Documentation/Nightly/Modules/Segmentations"
m (Text replacement - "\[https:\/\/(?:www|wiki).?slicer.org\/slicerWiki\/index.php\?title=([^ ]+)([^]]*)]" to "[https://www.slicer.org/wiki/$1$2]") |
|||
| (13 intermediate revisions by 4 users not shown) | |||
| Line 10: | Line 10: | ||
This work is part of the SparKit project, funded by An Applied Cancer Research Unit of Cancer Care Ontario with funds provided by the Ministry of Health and Long-Term Care and the Ontario Consortium for Adaptive Interventions in Radiation Oncology (OCAIRO) to provide free, open-source toolset for radiotherapy and related image-guided interventions.<br> | This work is part of the SparKit project, funded by An Applied Cancer Research Unit of Cancer Care Ontario with funds provided by the Ministry of Health and Long-Term Care and the Ontario Consortium for Adaptive Interventions in Radiation Oncology (OCAIRO) to provide free, open-source toolset for radiotherapy and related image-guided interventions.<br> | ||
Author: Csaba Pinter (PerkLab, Queen's University)<br> | Author: Csaba Pinter (PerkLab, Queen's University)<br> | ||
| − | Contact: Csaba Pinter, <email>csaba | + | Contact: Csaba Pinter, <email>pinter.csaba@gmail.com</email><br> |
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
{{documentation/{{documentation/version}}/module-introduction-logo-gallery | {{documentation/{{documentation/version}}/module-introduction-logo-gallery | ||
| Line 41: | Line 41: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Tutorials}} | {{documentation/{{documentation/version}}/module-section|Tutorials}} | ||
| − | * [ | + | |
| + | * [[Documentation/{{documentation/version}}/Training#Slicer4_Image_Segmentation|Segmentation tutorials]] | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|How to}} | {{documentation/{{documentation/version}}/module-section|How to}} | ||
| − | ; | + | ; Edit segmentation |
| − | : | + | : Segment structures in the '''[{{documentation/{{documentation/version}}/slicer-manual-base-url}}/user_guide/module_segmenteditor.html Segment Editor]''' module |
| − | ; | + | ; ''Import labelmap'' volume or ''model'' into segmentation |
| − | + | # In Segmentations module | |
| − | : | + | #* Expand Export/import models and labelmaps section at the bottom of the Segmentations module (it is collapsed by default) |
| − | + | #* Select Import, import type, then choose imported node: labelmap, model, or folder containing models | |
| + | #* Click Import | ||
| + | # In Data module (Subject hierarchy tab) | ||
| + | #* Right-click model or model hierarchy node | ||
| + | #* Select 'Convert model to segmentation node' | ||
| − | ; Export labelmap or surface model | + | ; ''Export labelmap'' or surface ''model'' from segmentation (for saving to STL, editing, etc). |
| − | + | # Expand the Export/import segments section at the bottom of the Segmentations module (it is collapsed by default). Select output type, then click Export. If exporting to an existing node is needed, it can be selected as Output node. If only a subset of the segments are needed, then open the Advanced section, where this selection can be changed, along with the reference volume. | |
| − | + | # Switch to the Data module and make sure the Subject hierarchy tree is selected. Right-click the segmentation node, then select one of the Export functions | |
: To save exported result to file, use the Save data dialog (left side of top toolbar). To save to STL, change the File Format selector to STL in the rows of the models to be saved. | : To save exported result to file, use the Save data dialog (left side of top toolbar). To save to STL, change the File Format selector to STL in the rows of the models to be saved. | ||
| Line 65: | Line 70: | ||
; Change display mode of segments | ; Change display mode of segments | ||
| − | + | # By long-pressing the eye icon in the segments table the display options are shown and the different display modes can be turned on or off | |
| − | + | # In Data module's Subject hierarchy, right-clicking the visibility eye icon next to the segmentation node or its segments also offers advanced display options | |
; Managing segmentations in python | ; Managing segmentations in python | ||
| − | : See [https://www.slicer.org/ | + | : See [https://www.slicer.org/wiki/Documentation/Nightly/ScriptRepository#Segmentations Segmentations section] in the script in repository |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 117: | Line 122: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Similar Modules}} | {{documentation/{{documentation/version}}/module-section|Similar Modules}} | ||
| − | * [ | + | * [http://slicer.readthedocs.io/en/latest/user_guide/module_segmenteditor.html Segment Editor] module is used to edit Segmentation nodes |
| − | * This module and infrastructure is a successor of the [[Documentation/4.4/Modules/Contours|Contours]] module and mechanism, which was the first implementation for this task, but its design was | + | * This module and infrastructure is a successor of the [[Documentation/4.4/Modules/Contours|Contours]] module and mechanism, which was the first implementation for this task, but its design was overgrown by the new features and needs, and became inconvenient and unstable. Further information about the Segmentation effort on the related [https://github.com/SlicerRt/SlicerRT/wiki/Segmentation design wiki page], the [https://www.slicer.org/wiki/Documentation/Labs/Segmentations labs wiki page], and the links pointing from these pages. |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|References}} | {{documentation/{{documentation/version}}/module-section|References}} | ||
* Past references | * Past references | ||
| + | <!-- | ||
** Issue for Segmentations (complete Contours overhaul, delivery June 2015) [https://www.assembla.com/spaces/slicerrt/tickets/626 here] | ** Issue for Segmentations (complete Contours overhaul, delivery June 2015) [https://www.assembla.com/spaces/slicerrt/tickets/626 here] | ||
** Issue for original Contours effort on SlicerRT Assembla ticketing system [https://www.assembla.com/spaces/slicerrt/tickets/137 here] | ** Issue for original Contours effort on SlicerRT Assembla ticketing system [https://www.assembla.com/spaces/slicerrt/tickets/137 here] | ||
| − | ** Development notes [https:// | + | --> |
| + | ** Development notes [https://github.com/SlicerRt/SlicerRT/wiki/Segmentation wiki page] | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Information for Developers}} | {{documentation/{{documentation/version}}/module-section|Information for Developers}} | ||
| − | * [ | + | * [https://github.com/Slicer/Slicer/tree/master/Libs/vtkSegmentationCore vtkSegmentationCore on GitHub] (within Slicer) |
| − | * Numerous scripting examples are listed in the [ | + | * [https://github.com/Slicer/Slicer/tree/master/Modules/Loadable/Segmentations Segmentations Slicer module on GitHub] |
| + | * [[Documentation/Labs/Segmentations|Labs page]] | ||
| + | * Numerous scripting examples are listed in the [[Documentation/Nightly/ScriptRepository|Script repository]] page | ||
* How to create new representation or converter - TODO | * How to create new representation or converter - TODO | ||
Revision as of 14:39, 27 November 2019
Home < Documentation < Nightly < Modules < Segmentations
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work is part of the SparKit project, funded by An Applied Cancer Research Unit of Cancer Care Ontario with funds provided by the Ministry of Health and Long-Term Care and the Ontario Consortium for Adaptive Interventions in Radiation Oncology (OCAIRO) to provide free, open-source toolset for radiotherapy and related image-guided interventions. | |||||||
|
Module Description
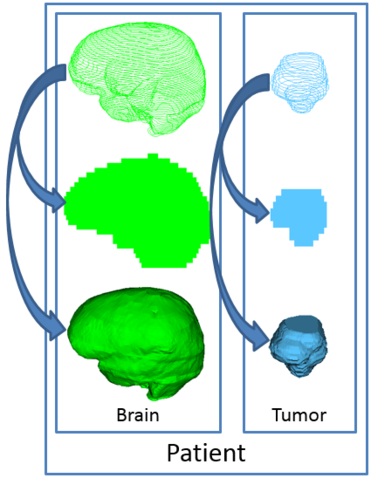
The Segmentations module manages segmentations. Each segmentation can contain multiple segments, which correspond to one structure or ROI. Each segment can contain multiple data representations for the same structure, and the module supports automatic conversion between these representations (the default ones are: planar contour, binary labelmap, closed surface model), as well as advanced display settings and import/export features.
Motivation, features, and details of the infrastructure are explained in these slides (PDF).
Note: It is important to remember that segmentations are not labelmaps, i.e. they are not one 3D volume containing color indices in a color table. There is a legacy labelmap support that temporarilymerges the segments into an old-school labelmap, but that feature will be removed, and under the hood the segmentation node is structured as explained above.
Use Cases
- 3D printing: Convert labelmaps to surface models and export them to STL
- Contour analysis: Convert RT planar contours to labelmap for per-voxel analysis
- Nice and flexible visualization: Show any structure as 3D surface, and line intersections in 2D views; Show surface and labelmap simultaneously
- Organize segmented structures: Each segmentation contains multiple "segments"
Tutorials
How to
- Edit segmentation
- Segment structures in the Segment Editor module
- Import labelmap volume or model into segmentation
- In Segmentations module
- Expand Export/import models and labelmaps section at the bottom of the Segmentations module (it is collapsed by default)
- Select Import, import type, then choose imported node: labelmap, model, or folder containing models
- Click Import
- In Data module (Subject hierarchy tab)
- Right-click model or model hierarchy node
- Select 'Convert model to segmentation node'
- Export labelmap or surface model from segmentation (for saving to STL, editing, etc).
- Expand the Export/import segments section at the bottom of the Segmentations module (it is collapsed by default). Select output type, then click Export. If exporting to an existing node is needed, it can be selected as Output node. If only a subset of the segments are needed, then open the Advanced section, where this selection can be changed, along with the reference volume.
- Switch to the Data module and make sure the Subject hierarchy tree is selected. Right-click the segmentation node, then select one of the Export functions
- To save exported result to file, use the Save data dialog (left side of top toolbar). To save to STL, change the File Format selector to STL in the rows of the models to be saved.
- Create new representation in segmentation (conversion)
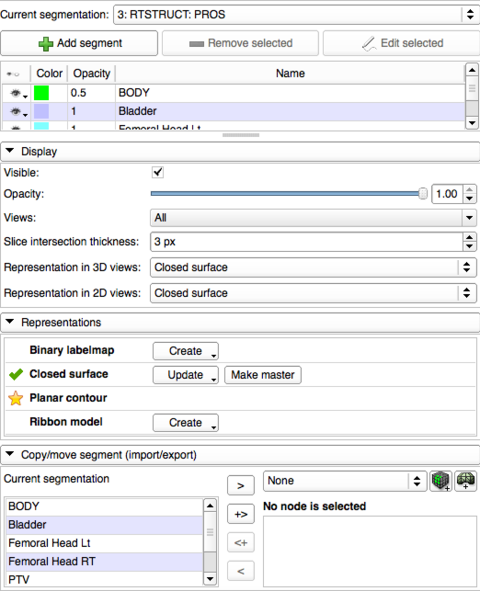
- The supported representations are listed as rows in the Representations section. The already existing representations have a green tick, the master representation (that is the source of all conversions and the representation that can be edited) a gold star
- To create a representation that is missing, click on the Create button in its row. If custom conversion path or parameters are to be used (reference grid for labelmaps, smoothing for surfaces, etc.), long-press the button and choose "Advanced create...". In the appearing window the available conversion paths are listed in a list, ordered by estimated time cost. After selecting the desired path, the involved parameters are listed in the bottom section of the window
- To re-convert an existing representation (to use different conversion path or parameters), click the Update buitton in their row
- Change display mode of segments
- By long-pressing the eye icon in the segments table the display options are shown and the different display modes can be turned on or off
- In Data module's Subject hierarchy, right-clicking the visibility eye icon next to the segmentation node or its segments also offers advanced display options
- Managing segmentations in python
- See Segmentations section in the script in repository
Panels and their use
- Segments table
- Add/remove segments
- Edit selected: takes user to Segment Editor module
- Set visibility and per-segment display settings, opacity, color, segment name
- Display
- Segmentations-wide display settings (not per-segment!): visibility, opacity (will be multiplied with per-segment opacity for display)
- Views: Individual views to show the active segmentation in
- Slice intersection thickness
- Representation in 3D/2D views: The representation to be shown in the 3D and 2D views. Useful if there are multiple representations available, for example if we want to show the closed surface in the 3D view but the labelmap in the slice views
- Representations
- List of supported representations and related operations
- The already existing representations have a green tick, the master representation (that is the source of all conversions and the representation that can be edited) a gold star
- The buttons in each row can be used to create, remove, update a representation
- Advanced conversion is possible (to use the non-default path or conversion parameters) by long-pressing the Create or Update button
- Existing representations can be made master by clicking Make master. The master representation is used as source for conversions, it is the one that can be edited, and saved to disk
- Copy/move (import/export)
- Left panel lists the segments in the active segmentation
- Right panel shows the external data container
- The arrow buttons van be used to copy (with plus sign) or move (no plus sign) segments between the segmentation and the external node
- New labelmap or model can be created by clicking the appropriate button on the top of the right panel
- Multiple segments can be exported into a labelmap. In case of overlapping segments, the subsequent segments will overwrite the previous ones!
Subject hierarchy support
- Segmentations are shown in subject hierarchy as any other node, with the exception that the contained segments are in a "virtual branch".
- The segments can be moved between segmentations, but drag&drop to anywhere other than under another segmentation is not allowed
- Special subject hierarchy features for segmentations
- Create representation: Create the chosen representation using the default path
- Special subject hierarchy features for segments
- Show only this segment: Useful if only one segment needs to be shown and there are many, so clicking the eye buttons woud take a long time
- Show all segments
DICOM export
- The master representation is used when exporting into DICOM
- RT structure set export is available if SlicerRT is installed
- DICOM SegObj export - coming soon!
Similar Modules
- Segment Editor module is used to edit Segmentation nodes
- This module and infrastructure is a successor of the Contours module and mechanism, which was the first implementation for this task, but its design was overgrown by the new features and needs, and became inconvenient and unstable. Further information about the Segmentation effort on the related design wiki page, the labs wiki page, and the links pointing from these pages.
References
- Past references
- Development notes wiki page
Information for Developers
- vtkSegmentationCore on GitHub (within Slicer)
- Segmentations Slicer module on GitHub
- Labs page
- Numerous scripting examples are listed in the Script repository page
- How to create new representation or converter - TODO
FAQ
- The closed surface generated from the RT Structure Set contour points is flawed
- Please report the issue first so that we can fix the conversion.
- Use advanced conversion and choose a path through Ribbon model representation. SlicerRT extension needs to be installed for this.