Difference between revisions of "Modules:FourDAnalysis-Documentation-3.5"
| Line 49: | Line 49: | ||
== Development == | == Development == | ||
===Installation=== | ===Installation=== | ||
| − | + | Although the module's source directory is in the trunk repository, it is not included in the CMake configuration. | |
| + | To use 4D Analysis Module, you need edit Slicer3/Modules/CMakeLists.txt and build the module. | ||
| − | + | (Supposing you have your local copy of 3D SLicer under <working directory>, in the following instruction) | |
| − | |||
| − | + | ====1. Edit Slicer3/Modules/CMakeLists.txt==== | |
| − | + | In '''subdirs''' section, insert 'FourDAnalysis' as: | |
| + | subdirs( | ||
| + | MRIBiasFieldCorrection | ||
| + | |||
| + | ... | ||
| + | |||
| + | FastMarchingSegmentation | ||
| + | FourDImage | ||
| + | FourDAnalysis # <- Here!! | ||
| + | ExtractSubvolumeROI | ||
| + | |||
| + | ... | ||
| + | ) | ||
| − | + | ====2. Build the module==== | |
| − | + | In Linux or Mac OS X, | |
| − | + | cd <working directory>/Slicer3-build/Modules | |
| − | + | make | |
| − | |||
| − | + | In Windows, open the project file in <working directory>/Slicer3-build/Modules, and build all. | |
| − | |||
| − | |||
| − | If the module | + | ====3. Check the module==== |
| + | If the module has been built successfully, it shows up in the '4D' section in the module selection list. | ||
Revision as of 01:07, 21 August 2009
Home < Modules:FourDAnalysis-Documentation-3.5Return to Slicer 3.5 Documentation
Module Name
4D Analysis Module
General Information
Module Type & Category
Type: Interactive
Category: 4D
Authors, Collaborators & Contact
- Junichi Tokuda, Ph.D., Brigham and Women's Hospital
- Hiroto Hatabu, M.D., Ph.D., Brigham and Women's Hospital
- Contact: Junichi Tokuda, tokuda at bwh.harvard.edu
Module Description
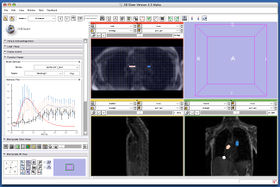
The 4D Analysis Module allows you to load, visualize and analyze a time-series of 3D images (4D image) on 3D Slicer. Currently, the module offers the following functions:
- 4D image loader. The module can load up a series of 3D images from a specified directory. It analyzes DICOM header of the files in the directory and split them into time-frames, even if all frames have the same series number (some MRI scanners generates time-series images without incrementing series number, resulting a large number (typically several thousands) of 2D image files output in a single directory). The module can also read a directory, which contains NRRD files.
- Time scroll. The module provides a scroll-bar interface to scroll the frame in time-direction. It allows you to scroll the frame for foreground and background screens independently to compare two images at the different time points.
- Intensity plot. The module can draw intensity curve at the specified region of interest.
- Series registration. The module provides a user-interface to perform image registration for each frame with a key frame. This function is useful if you want to perform intensity curve analysis on a series of images of an organ moving through the frames.
Usage
Examples, Use Cases & Tutorials
- Note use cases for which this module is especially appropriate, and/or link to examples.
- Link to examples of the module's use
- Link to any existing tutorials
Quick Tour of Features and Use
List all the panels in your interface, their features, what they mean, and how to use them. For instance:
- Input panel:
- Parameters panel:
- Output panel:
- Viewing panel:
Development
Installation
Although the module's source directory is in the trunk repository, it is not included in the CMake configuration. To use 4D Analysis Module, you need edit Slicer3/Modules/CMakeLists.txt and build the module.
(Supposing you have your local copy of 3D SLicer under <working directory>, in the following instruction)
1. Edit Slicer3/Modules/CMakeLists.txt
In subdirs section, insert 'FourDAnalysis' as:
subdirs( MRIBiasFieldCorrection ... FastMarchingSegmentation FourDImage FourDAnalysis # <- Here!! ExtractSubvolumeROI ... )
2. Build the module
In Linux or Mac OS X,
cd <working directory>/Slicer3-build/Modules make
In Windows, open the project file in <working directory>/Slicer3-build/Modules, and build all.
3. Check the module
If the module has been built successfully, it shows up in the '4D' section in the module selection list.
Dependencies
The intensity curve analysis functions depend on SciPy package. Please turn on SciPy option when you build your 3D Slicer.
Known bugs
Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Customize following links for your module.
Links to documentation generated by doxygen.
More Information
Acknowledgment
This work is supported by NIH (5R21CA116271: Dynamic contrast-enhanced MRI of pulmonary nodule at 3T, PI: Hiroto Hatabu), NA-MIC, and NCIGT.