Modules:PolyDataToLabelmap-Documentation-3.6
Return to Slicer 3.6 Documentation
Module Name
PolyData to LabelMap
General Information
Module Type & Category
Type: CLI
Category: Surface Models
Authors, Collaborators & Contact
- Author: Xiaodong Tao and Nicole Aucoin
- Contact: taox at research.ge.com
Module Description
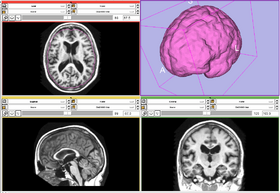
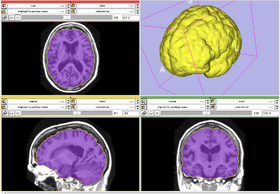
This module creates a label map of the interior of an input model within a reference volume. The module first label voxels that intersects the surface as foreground, then use a flood fill to label all interior.
Usage
Examples, Use Cases & Tutorials
- This module may be thought of as the inverse of Marching Cubes.
- A command line example:
./PolyDataToLabelmap --distance 1.0
InputReferenceVolume.nrrd
InputSurfaceModel.vtp
OutputLabelMap.nrrd
Quick Tour of Features and Use
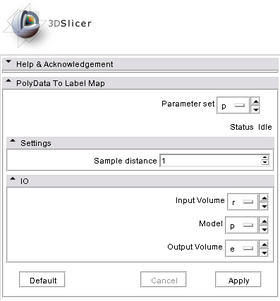
- Input/Output panels:
This module takes three input: Input Volume is the reference volume. Model is the surface model from whose interior is to be labeled as foreground, and Output Volume is the output label map. Output Volume has the same image parameters (size, resolution, orientation, etc) as Input Volume.
- Parameters panel:
Since surface can be very sparsely triangulated, this module first resamples the input surface model in an appropriate resolution. The parameter Sample Distance controls how fine the surface is sampled. The ballpark value for this parameter is the smallest size of voxels (in mm) in Input Volume.
Development
Dependencies
This module depends only on the core "Volumes" module of slice for data IO.
Known bugs
None.
Usability issues
When the object has a geometry such that the centroid of all vertices lie outside of the object, the module will fail to generate the correct label for the object.
Source code & documentation
Source Code: PolyDataToLabelmap.cxx
XML Description: PolyDataToLabelmap.xml
Test: PolyDataToLabelmapTest.cxx
Usage:
./PolyDataToLabelmap --help
USAGE:
./PolyDataToLabelmap [--returnparameterfile <std::string>]
[--processinformationaddress <std::string>]
[--xml] [--echo] [--distance <float>] [--]
[--version] [-h] <std::string> <std::string>
<std::string>
Where:
--returnparameterfile <std::string>
Filename in which to write simple return parameters (int, float,
int-vector, etc.) as opposed to bulk return parameters (image,
geometry, transform, measurement, table).
--processinformationaddress <std::string>
Address of a structure to store process information (progress, abort,
etc.). (default: 0)
--xml
Produce xml description of command line arguments (default: 0)
--echo
Echo the command line arguments (default: 0)
--distance <float>
Sample distance (default: 1)
--, --ignore_rest
Ignores the rest of the labeled arguments following this flag.
--version
Displays version information and exits.
-h, --help
Displays usage information and exits.
<std::string>
(required) Input volume
<std::string>
(required) Model
<std::string>
(required) The label volume
Description: Intersects an input model with an reference volume and
produces an output label map.
Author(s): Nicole Aucoin BWH, Xiaodong Tao, GE
Acknowledgements: This work is part of the National Alliance for Medical
Image Computing (NAMIC), funded by the National Institutes of Health
through the NIH Roadmap for Medical Research, Grant U54 EB005149.
More Information
Acknowledgment
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on the National Centers for Biomedical Computing can be obtained from National Centers for Biomedical Computing.
OASIS datasets were used to generate images on this page.
References
Note: this CLI module requires .vtp format input (this is the "new" VTK data format that uses XML encoding).
If you need to do a batch conversion of .vtk to .vtp, you may want to use File:Vtk2xml.zip originally by Prabhu Ramachandran <prabhu@aero.iitm.ernet.in> and enhanced for this purpose by András Jakab MD <jakaba@med.unideb.hu>
Another option, from Thomas Ballinger <tomb@bwh.harvard.edu> is available here.