Difference between revisions of "Slicer 4.0:Training"
From Slicer Wiki
(Create 4.0 tutorials page) |
|||
| Line 1: | Line 1: | ||
=Slicer 4.0 Tutorials= | =Slicer 4.0 Tutorials= | ||
| + | |||
| + | |||
| + | <br> | ||
| + | |||
| + | {| border="1" cellpadding="3" width="1000px" | ||
| + | |- style="background:#CCFF99; color:black; font-size:130%" align="center" | ||
| + | |style="width:100px"|'''Category''' | ||
| + | |style="width:350px"|'''Steps Taken''' | ||
| + | |style="width:350px"|'''Problems''' | ||
| + | |style="width:250px"|'''Image''' | ||
| + | |- | ||
| + | | id="Slicer3MinuteTutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Core tutorial 1''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top"| '''[[Media:Slicer3Minute_SoniaPujol_3.6.1.pdf | Slicer3Minute Tutorial]]'''<br><br><br>1)open slicer. 2)opened data set. 3)rotated model. 4)clicked slice visibility icon (red). 5)turn skin opacity to 0.0. 6) clicked slice visibility icon (green). 7)clip the scull and bone exposing the white matter tissue. 8) move through the scan (green). 9)take visibility off white matter. 10) zoom in and out. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" | '''[http://www.slicer.org/slicerWiki/images/5/51/Slicer3MinuteDataset.zip Slicer3Minute Data]'''<br><br> step 2) picture for tutorial is out of date. | ||
| + | | style="background:#FFFFCC; color:black" align="center"| [[Image:Slicer3Minute_3.6RC2.png |250px]] | ||
| + | |- | ||
| + | | id="Slicer3VisualizationTutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Core''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top"| '''[[media:Slicer3_DataLoadingAndVisualization_UCSF2010_SoniaPujol.pdf | Slicer3Visualization Tutorial]]'''<br><br><br>The Slicer3Visualization tutorial guides through 3D data loading and visualization in Slicer3.6.<br><br>'''Audience:''' All beginners including clinicians, scientists, engineers and programmers. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" | '''[http://www.slicer.org/slicerWiki/images/6/61/Slicer3VisualizationDataset.zip Slicer3Visualization Data]'''<br><br>The Slicer3VisualizationDataset contains two MR scans of the brain, a pre-computed labelmap and 3D reconstructions of the anatomy. | ||
| + | | style="background:#FFFFCC; color:black" align="center"| [[Image:VisualizationTutorial 3.6RC3.png|250px]] | ||
| + | |- | ||
| + | | id="ProgrammingInSlicer3Tutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Core''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top"|''' [[media:ProgrammingIntoSlicer3.6_SoniaPujol.pdf |Programming in Slicer3 Tutorial]]'''<br><br><br>The Programming in Slicer3 tutorial is an introduction to the integration of C++ stand-alone programs outside of the Slicer3 source tree.<br><br>'''Audience:''' Programmers and Engineers. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" | [[Media:HelloWorld_Plugin.zip| '''HelloWorld Plugin''']]<br><br> The HelloWorld tutorial dataset contains an MR scan of the brain and pre-computed xml and C++ files for integrating the Hello Python plug-in to Slicer3. | ||
| + | | style="background:#FFFFCC; color:black" align="center"| [[Image:ProgrammingCourse_Logo.PNG|250px|Programming]] | ||
| + | |- | ||
| + | | id="HelloPythonTutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Core''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top"|''' [[media:ProgrammingIntoSlicer3.6.1_HelloPython_MICCAI2010_SoniaPujol.pdf |Hello Python Tutorial]]''' <br><br>The Hello Python tutorial is an introduction to the integration of Python stand-alone programs outside of the Slicer3 source tree.<br><br>'''Audience:''' Programmers and Engineers. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" | [[Media:HelloPython.zip | '''HelloPython Plugin''']]<br><br> The HelloPython tutorial dataset contains an MR scan of the brain and pre-computed xml and Python code for integrating the Hello Python plug-in to Slicer3. | ||
| + | | style="background:#FFFFCC; color:black" align="center"| [[Image:HelloPythonTutorial.PNG|250px|Programming]] | ||
| + | |- | ||
| + | | id="InteractiveEditor" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Core''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top"|'''[[media:InteractiveEditorTutorial_Slicer3.6-SoniaPujol.pdf |Interactive Editor]] '''<br><br><br>Shows how to use the interactive editing tools in Slicer. <br>'''Audience:''' All users and developers. | ||
| + | | style="background:#FFFFCC; color:black" valign="top"|'''[http://www.slicer.org/slicerWiki/index.php/File:EditorTutorialDataset.zip Editor Data]'''<br><br>This dataset contains a MR dataset of the brain. | ||
| + | | style="background:#FFFFCC; color:black" align="center"|[[Image:InteractiveEditor.png|250px]] | ||
| + | |- | ||
| + | | id="ManualRegistration" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Core''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top"|'''[[media:ManualRegistration_Slicer3.6.pdf | Manual Registration]] ''' <br><br><br> Shows how to manually/interactively align two images in Slicer3.6 <br><br>'''Audience:''' First time & early users. | ||
| + | | style="background:#FFFFCC; color:black" valign="top"|'''[http://www.slicer.org/slicerWiki/images/8/88/Slicer3_Tutorial_ManualRegistration_ExampleDataset.zip Manual Registration Data]'''<br><br>This dataset contains two brain MRI of a single subject, obtained in different orientations. | ||
| + | | style="background:#FFFFCC; color:black" align="center"|[[Image:Slicer3_ManualRegistrationTutorial.gif|250px]] | ||
| + | |- | ||
| + | | id="DiffusionMRITutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top"|'''[[Media:DiffusionMRITutorial_UCSF2010_SoniaPujol.pdf | Diffusion MRI tutorial]]''' <br><br><br>This tutorial guides you through the process of loading diffusion MR data, estimating diffusion tensors, and performing tractography of white matter bundles. <br><br>'''Audience:''' All users and developers. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |'''[http://www.slicer.org/slicerWiki/images/c/cf/DiffusionDataset.zip Diffusion Data]''' | ||
| + | | style="background:#FFFFCC; color:black" align="center"|[[Image:cc.PNG |250px]] | ||
| + | |- | ||
| + | | id="ChangeTrackerTutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top"|'''[[media:Slicer3.6-ChangeTrackerTutorial.pdf | Change Tracker Tutorial]]''' <br><br><br>This tutorial describes the use of ChangeTracker module to detect changes in tumor volume from two MRI scans. <br><br>'''Audience:''' All users interested in longitudinal analysis of pathology. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |Training Data download is integrated with the ChangeTracker module (see Tutorial) | ||
| + | | style="background:#FFFFCC; color:black" align="center"|[[Image:Slicer3.4.1-ChangeTracker.jpg|250px]] | ||
| + | |- | ||
| + | | id="FreeSurferCourse" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top"|'''[[media:FreeSurferCourse_Slicer3.6_SoniaPujol.pdf | FreeSurfer Course ]]''' The FreeSurfer dataset contains an MR scan of the brain and pre-computed FreeSurfer segmentation and cortical surface reconstructions. | ||
| + | <br><br>'''Audience:''' All users | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |'''[http://www.na-mic.org/Wiki/index.php/File:FreeSurferTutorialData.zip FreeSurfer tutorial data '''] | ||
| + | | style="background:#FFFFCC; color:black" align="center|[[Image:FreeSurferTutorial.PNG|250px]] | ||
| + | |- | ||
| + | | id="NeurosurgicalPlanningTutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top"| [[media:SlicerNeurosurgeryTutorial-3.6.1.pdf| '''Neurosurgical Planning Tutorial''']] <br><br> This tutorial takes the trainee through a complete workup for neurosurgical patient-specific mapping. Also see this tutorial for information on how to use Slicer's affine registration, simple region growing, model maker and tractography modules.<br> <br>'''Audience:''' All users interested in image-guided therapy. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" | [[Media:Slicer-tutorial-neurosurgery_slicer3.6.1.zip| '''Neurosurgical Planning Data ''']] | ||
| + | | style="background:#FFFFCC; color:black" align="center"| [[Image:NeurosurgicalPlanningOverview.png|250px]] | ||
| + | |- | ||
| + | | id="PETCTSUVTutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top"| [http://www.na-mic.org/Wiki/index.php/File:PETCTTutorial.pptx '''PET/CT SUV Tutorial'''] <br><br> This tutorial takes the trainee through the computation of SUV body weight on a baseline and followup study. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" | [http://wiki-na-mic.org/Wiki/images/7/73/PETCTFusion-Tutorial-Data.zip '''PET/CT Data '''] | ||
| + | | style="background:#FFFFCC; color:black" align="center"| [[Image:PETCTFusionBig.png|250px]] | ||
| + | |- | ||
| + | | id="EMSegmenterSimpleMode" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top"| [[media:EMSegmenterTutorialSimpleMode-Slicer-3.6.3-1.pdf | '''EMSegmenter Simple Mode ''']] <br><br> This tutorial takes the trainee through the segmentation of a MRI Human Brain without to adjust any parameters. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" | [http://www.slicer.org/slicerWiki/images/c/cd/MRIHumanBrain_T1_aligned.nrrd '''Human Brain T1 Data'''] | ||
| + | | style="background:#FFFFCC; color:black" align="center"| [[Image:EMSegmenter-Simple-Mode.png|250px]] | ||
| + | |- | ||
| + | | id="EMSegmenterAdvancedMode" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top"| [[media:EMSegmenterTutorialAdvancedMode-Slicer-3.6.3-2.pdf | '''EMSegmenter Advanced Mode ''']] <br><br> This tutorial takes the trainee through the segmentation of a MRI Human Brain. The trainee will learn how to setup the EMSegmenter, this includes the creation of a task, the creation of an anatomical tree and adjusting the weights for the EM algorithm. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" | [http://www.slicer.org/slicerWiki/images/c/cd/MRIHumanBrain_T1_aligned.nrrd '''Human Brain T1 Data'''] | ||
| + | | style="background:#FFFFCC; color:black" align="center"| [[Image:EMSegmenter-Advanced-Mode.png|250px]] | ||
| + | |- | ||
| + | | id="OpenIGTLink" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top"|[[media:OpenIGTLinkTutorial_Slicer3.6.2_JunichiTokuda_Dec2010.pdf | '''OpenIGTLink''']] <br><br> This tutorial shows to connect an IGT device using the OpenIGTLink. | ||
| + | |style="background:#FFFFCC; color:black" valign="top" | [http://wiki.slicer.org/slicerWiki/index.php/Modules:OpenIGTLinkIF-3.6-Simulators'''OpenIGTLink Data Simulator'''] | ||
| + | | style="background:#FFFFCC; color:black" align="center"| [[Image:OpenIGT.PNG|250px]] | ||
| + | |- | ||
| + | | id="WhiteMatterExplorationForNeurosurgicalPlanning" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Workflow''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top"|[[media:WhiteMatterExplorationNeurosurgicalPlanning_JHU2011_SoniaPujol.pdf | '''White Matter Exploration for Neurosurgical Planning''']] <br><br> This [[White_Matter_Exploration_Tutorial | tutorial]] walks the user through a workflow for exploring white matter fibers surrounding a tumor using Diffusion Tensor Imaging (DTI) Tractography. | ||
| + | |style="background:#FFFFCC; color:black" valign="top" | [[media:WhiteMatterExplorationData.zip| White Matter Exploration dataset]] | ||
| + | | style="background:#FFFFCC; color:black" align="center"| [[Image:WhiteMatterExploration.PNG|250px]] | ||
| + | |- | ||
| + | |} | ||
Revision as of 15:25, 5 July 2011
Home < Slicer 4.0:TrainingSlicer 4.0 Tutorials
| Category | Steps Taken | Problems | Image |
| Core tutorial 1 | Slicer3Minute Tutorial 1)open slicer. 2)opened data set. 3)rotated model. 4)clicked slice visibility icon (red). 5)turn skin opacity to 0.0. 6) clicked slice visibility icon (green). 7)clip the scull and bone exposing the white matter tissue. 8) move through the scan (green). 9)take visibility off white matter. 10) zoom in and out. |
Slicer3Minute Data step 2) picture for tutorial is out of date. |
|
| Core | Slicer3Visualization Tutorial The Slicer3Visualization tutorial guides through 3D data loading and visualization in Slicer3.6. Audience: All beginners including clinicians, scientists, engineers and programmers. |
Slicer3Visualization Data The Slicer3VisualizationDataset contains two MR scans of the brain, a pre-computed labelmap and 3D reconstructions of the anatomy. |
|
| Core | Programming in Slicer3 Tutorial The Programming in Slicer3 tutorial is an introduction to the integration of C++ stand-alone programs outside of the Slicer3 source tree. Audience: Programmers and Engineers. |
HelloWorld Plugin The HelloWorld tutorial dataset contains an MR scan of the brain and pre-computed xml and C++ files for integrating the Hello Python plug-in to Slicer3. |
|
| Core | Hello Python Tutorial The Hello Python tutorial is an introduction to the integration of Python stand-alone programs outside of the Slicer3 source tree. Audience: Programmers and Engineers. |
HelloPython Plugin The HelloPython tutorial dataset contains an MR scan of the brain and pre-computed xml and Python code for integrating the Hello Python plug-in to Slicer3. |
|
| Core | Interactive Editor Shows how to use the interactive editing tools in Slicer. Audience: All users and developers. |
Editor Data This dataset contains a MR dataset of the brain. |
|
| Core | Manual Registration Shows how to manually/interactively align two images in Slicer3.6 Audience: First time & early users. |
Manual Registration Data This dataset contains two brain MRI of a single subject, obtained in different orientations. |
|
| Specialized | Diffusion MRI tutorial This tutorial guides you through the process of loading diffusion MR data, estimating diffusion tensors, and performing tractography of white matter bundles. Audience: All users and developers. |
Diffusion Data | 
|
| Specialized | Change Tracker Tutorial This tutorial describes the use of ChangeTracker module to detect changes in tumor volume from two MRI scans. Audience: All users interested in longitudinal analysis of pathology. |
Training Data download is integrated with the ChangeTracker module (see Tutorial) | |
| Specialized | FreeSurfer Course The FreeSurfer dataset contains an MR scan of the brain and pre-computed FreeSurfer segmentation and cortical surface reconstructions.
|
FreeSurfer tutorial data | 
|
| Specialized | Neurosurgical Planning Tutorial This tutorial takes the trainee through a complete workup for neurosurgical patient-specific mapping. Also see this tutorial for information on how to use Slicer's affine registration, simple region growing, model maker and tractography modules. Audience: All users interested in image-guided therapy. |
Neurosurgical Planning Data | 
|
| Specialized | PET/CT SUV Tutorial This tutorial takes the trainee through the computation of SUV body weight on a baseline and followup study. |
PET/CT Data | 
|
| Specialized | EMSegmenter Simple Mode This tutorial takes the trainee through the segmentation of a MRI Human Brain without to adjust any parameters. |
Human Brain T1 Data | 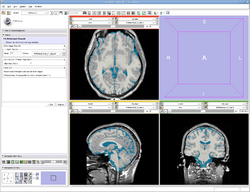
|
| Specialized | EMSegmenter Advanced Mode This tutorial takes the trainee through the segmentation of a MRI Human Brain. The trainee will learn how to setup the EMSegmenter, this includes the creation of a task, the creation of an anatomical tree and adjusting the weights for the EM algorithm. |
Human Brain T1 Data | 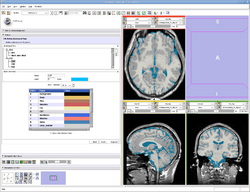
|
| Specialized | OpenIGTLink This tutorial shows to connect an IGT device using the OpenIGTLink. |
OpenIGTLink Data Simulator | 
|
| Workflow | White Matter Exploration for Neurosurgical Planning This tutorial walks the user through a workflow for exploring white matter fibers surrounding a tumor using Diffusion Tensor Imaging (DTI) Tractography. |
White Matter Exploration dataset | 
|