Slicer3:XNAT
From Slicer Wiki
Home < Slicer3:XNAT
Contents
Background
- XNAT is available at http://www.xnat.org
- Sample XNAT sites:
- http://www.oasis-brains.org
- http://iaclin2.wustl.edu/mbdr
- http://yaz.bwh.harvard.edu:8080/xnat/ (not working: being used to debug Rocks-based install)
- http://yaz.bwh.harvard.edu:8080/xnat2/ (does work; manual install)
- http://sandbox.xnat.org/
Existing Slicer3 Interface
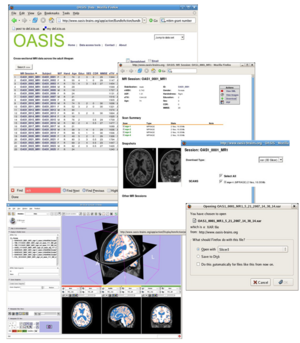
Basic steps:
- Browse to the desired image data set
- Select Download Images
- Pick which images you want
- Select the filetype as "xar (3D Slicer)"
- When the browser asks what to do with the file, select Slicer3 as the application (browse to where the program is installed).
- Once downloaded, Slicer will look in the file and offer to load the images.
Planned improvements:
- Fundamental goals for this project:
- is to be able to upload raw data into the database (core xnat feature)
- download selected datasets to slicer (see above)
- run analysis routines in slicer
- upload the results linked back to original data
- Further nice-to-have goals:
- use XNAT as an optional back end for the integration of BatchMake for population processing and parameter space exploration
- upload analysis results (numerical values) to database for plotting and other analysis
Installation
Options:
- instructions on http://xnat.org
- use the BIRN ROCKS installer (contact BIRN-CC via http://www.nbirn.net)
- use the xnat installer script from Gabriele Fariello (fariello at fas.harvard.edu)
- this works for some linux distributions (Red Hat and probably Ubuntu)
- you must have Sun Java installed (not the gnu version)
- best to use this installer on a fresh machine
- apt-get of postgres may hang waiting for you to press 'Y'
Tests
Get the xnat webservices command line tools from http://www.xnat.org/tools.html#
ArcGet -host www.oasis-brains.org -u guest -p guest -s OAS1_0001_MR1
Further XCEDE Integration
User Requests
Potential users of this project, please list feature requests and/or proposed use scenarios in this section.
- How is patient deidentification handled in XNAT?