Difference between revisions of "Slicer3:Remote Data Handling"
| Line 31: | Line 31: | ||
[[image:DataLoadingCurrent.png]] | [[image:DataLoadingCurrent.png]] | ||
| − | + | = | |
| − | = Goal for how Slicer would upload/download from remote data stores = | + | = Goal for how Slicer would upload/download from remote data stores == |
Eventually, we would like to query web services, download data remotely or locally from the Application itself, and have the option of uplaoding to remote stores as well. A sketch of the architecture planned in a meeting (on 2/14/08 with Steve Pieper, Nicole Aucoin and Wendy Plesniak) is shown below, including: | Eventually, we would like to query web services, download data remotely or locally from the Application itself, and have the option of uplaoding to remote stores as well. A sketch of the architecture planned in a meeting (on 2/14/08 with Steve Pieper, Nicole Aucoin and Wendy Plesniak) is shown below, including: | ||
* a collection of vtkURIHandlers, | * a collection of vtkURIHandlers, | ||
| Line 42: | Line 42: | ||
[[image:DataLoadingTarget.png ]] | [[image:DataLoadingTarget.png ]] | ||
| − | = Two general use cases used to drive a first pass implementation = | + | == Two general use cases used to drive a first pass implementation == |
* '''First''', is loading a combined FIPS/FreeSurfer analysis, specified in an Xcede catalog (.xcat) file that contains uris pointing to remote datasets, and view this with Slicer's QueryAtlas. (Prior to fBIRN AHM in 2008, it was not possible to get an .xcat via the HID web GUI; our approach was to manually upload a test Xcede catalog file and its constituent datasets to SRB. A copy of the .xcat file was kept locally, and SRB was accessed for each uri listed in it.) | * '''First''', is loading a combined FIPS/FreeSurfer analysis, specified in an Xcede catalog (.xcat) file that contains uris pointing to remote datasets, and view this with Slicer's QueryAtlas. (Prior to fBIRN AHM in 2008, it was not possible to get an .xcat via the HID web GUI; our approach was to manually upload a test Xcede catalog file and its constituent datasets to SRB. A copy of the .xcat file was kept locally, and SRB was accessed for each uri listed in it.) | ||
| Line 51: | Line 51: | ||
[[image:DataLoadingStartPlan.png]] | [[image:DataLoadingStartPlan.png]] | ||
| − | = Current MRML, Logic and GUI Components = | + | == Current MRML, Logic and GUI Components == |
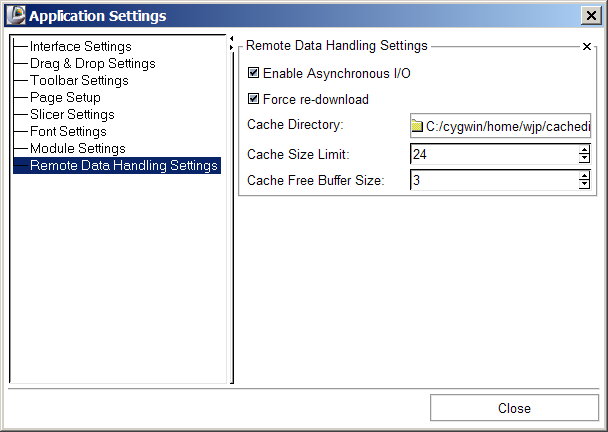
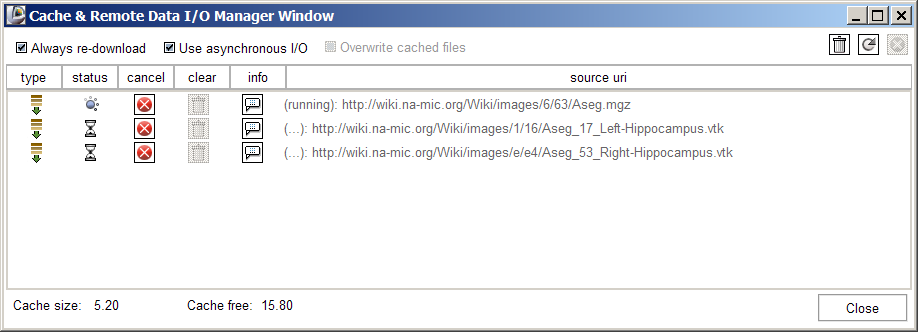
Slicer's Remote data handling architecture has been implemented to support remote download and upload of uris. Its components include a Cache Manager, a Data I/O Manager, and a URI Handler Collection in MRML; a Data I/O Manager Logic that engages the Application Logic's queueing and multi-threading mechanisms; and a Cache and Data I/O Manager GUI to provide feedback, information and control on the desktop. These components can also be configured through the Application Settings Interface, and these user preferences are saved in the Application Registry. Schematic and screenshots are shown below: | Slicer's Remote data handling architecture has been implemented to support remote download and upload of uris. Its components include a Cache Manager, a Data I/O Manager, and a URI Handler Collection in MRML; a Data I/O Manager Logic that engages the Application Logic's queueing and multi-threading mechanisms; and a Cache and Data I/O Manager GUI to provide feedback, information and control on the desktop. These components can also be configured through the Application Settings Interface, and these user preferences are saved in the Application Registry. Schematic and screenshots are shown below: | ||
| Line 82: | Line 82: | ||
* can XNAT-generated uris end with the filename.extension, so that firefox will know to save them to disk, rather than to interpret them as html? | * can XNAT-generated uris end with the filename.extension, so that firefox will know to save them to disk, rather than to interpret them as html? | ||
| − | = Implementation: new classes = | + | == Implementation: new classes == |
| − | ==MRML extensions== | + | ===MRML extensions=== |
'''MRML-specific implementations and extensions''' of the following classes: | '''MRML-specific implementations and extensions''' of the following classes: | ||
| Line 94: | Line 94: | ||
* vtkURIHandler | * vtkURIHandler | ||
| − | ==GUI extensions== | + | ===GUI extensions=== |
'''GUI implementations''' of the following classes: | '''GUI implementations''' of the following classes: | ||
| Line 101: | Line 101: | ||
* vtkSlicerPasswordPrompter | * vtkSlicerPasswordPrompter | ||
| − | ==Logic extensions== | + | ===Logic extensions=== |
* vtkDataIOManagerLogic | * vtkDataIOManagerLogic | ||
| − | ==Application Interface extensions== | + | ===Application Interface extensions=== |
* Set CacheDirectory ''default = Slicer3 temp dir'' | * Set CacheDirectory ''default = Slicer3 temp dir'' | ||
| Line 115: | Line 115: | ||
* Instance & Register URI Handlers (?) | * Instance & Register URI Handlers (?) | ||
| − | ==Important Notes== | + | ===Important Notes=== |
* MRML Scene file reading will always be synchronous. | * MRML Scene file reading will always be synchronous. | ||
* Read/write of individual datasets referenced in the scene should work with or without the asynchronous read/write turned on, and with or without the dataIOManager GUI interface. | * Read/write of individual datasets referenced in the scene should work with or without the asynchronous read/write turned on, and with or without the dataIOManager GUI interface. | ||
| − | = ITK-based mechanism handling remote data (for command line modules, batch processing, and grid processing) (Nicole) = | + | == ITK-based mechanism handling remote data (for command line modules, batch processing, and grid processing) (Nicole) == |
This one is tenatively on hold for now. | This one is tenatively on hold for now. | ||
| − | = Workflows to support = | + | == Workflows to support == |
The first goal is to figure out what workflows to support, and a good implementation approach. | The first goal is to figure out what workflows to support, and a good implementation approach. | ||
| Line 140: | Line 140: | ||
For now, we choose the first option. | For now, we choose the first option. | ||
| − | ==workflows:== | + | ===workflows:=== |
'''Possible workflow A''' | '''Possible workflow A''' | ||
| Line 155: | Line 155: | ||
In each workflow, the data gets saved to disk first and then loaded into StorageNode or uploaded to remote location from cache. | In each workflow, the data gets saved to disk first and then loaded into StorageNode or uploaded to remote location from cache. | ||
| − | == What data do we need in an .xcat file? == | + | === What data do we need in an .xcat file? === |
For the fBIRN QueryAtlas use case, we need a combination of '''FreeSurfer morphology analysis''' and a '''FIPS analysis''' of the same subject. With the combined data in Slicer, we can view activation overlays co-registered to and overlayed onto the high resolution structural MRI using the FIPS analysis, and determine the names of brain regions where activations occur using the co-registered morphology analysis. | For the fBIRN QueryAtlas use case, we need a combination of '''FreeSurfer morphology analysis''' and a '''FIPS analysis''' of the same subject. With the combined data in Slicer, we can view activation overlays co-registered to and overlayed onto the high resolution structural MRI using the FIPS analysis, and determine the names of brain regions where activations occur using the co-registered morphology analysis. | ||
The required analyses including all derived data are in two standard directory structures on local disk, and *hopefully* somewhere on the HID within a standard structure (check with Burak). These directory trees contain a LOT of files we don't need... Below are the files we *do* need for fBIRN QueryAtlas use case. | The required analyses including all derived data are in two standard directory structures on local disk, and *hopefully* somewhere on the HID within a standard structure (check with Burak). These directory trees contain a LOT of files we don't need... Below are the files we *do* need for fBIRN QueryAtlas use case. | ||
| − | ===FIPS analysis (.feat) directory and required data=== | + | ====FIPS analysis (.feat) directory and required data==== |
For instance, the FIPS output directory in our example dataset from Doug Greve at MGH is called sirp-hp65-stc-to7-gam.feat. Under this directory, QueryAtlas needs the following datasets: | For instance, the FIPS output directory in our example dataset from Doug Greve at MGH is called sirp-hp65-stc-to7-gam.feat. Under this directory, QueryAtlas needs the following datasets: | ||
* sirp-hp65-stc-to7-gam.feat/reg/example_func.nii | * sirp-hp65-stc-to7-gam.feat/reg/example_func.nii | ||
| Line 167: | Line 167: | ||
* sirp-hp65-stc-to7-gam.feat/design.gif (this image relates statistics files to experimental conditions) | * sirp-hp65-stc-to7-gam.feat/design.gif (this image relates statistics files to experimental conditions) | ||
| − | ===FreeSurfer analysis directory, and required data === | + | ====FreeSurfer analysis directory, and required data ==== |
For instance, the FreeSurfer morphology analysis directory in our example dataset from Doug Greve at MGH is called fbph2-000670986943. Under this directory, QueryAtlas needs the following datasets: | For instance, the FreeSurfer morphology analysis directory in our example dataset from Doug Greve at MGH is called fbph2-000670986943. Under this directory, QueryAtlas needs the following datasets: | ||
| Line 177: | Line 177: | ||
* fbph2-000670986943/label/rh.aparc.annot | * fbph2-000670986943/label/rh.aparc.annot | ||
| − | = What do we want HID webservices to provide? = | + | == What do we want HID webservices to provide? == |
* Question: are FIPS and FreeSurfer analyses (including QueryAtlas required files listed above) for subjects available on the HID yet? --Burak says not yet. | * Question: are FIPS and FreeSurfer analyses (including QueryAtlas required files listed above) for subjects available on the HID yet? --Burak says not yet. | ||
Revision as of 19:47, 2 April 2010
Home < Slicer3:Remote Data Handling
Contents
- 1 XNAT-related Slicer projects
- 1.1 Link to Fetch Medical Informatics (FetchMI) implementation 1 notes
- 1.2 Link to Fetch Medical Informatics (FetchMI) and RemoteIO profiling/refining notes
- 1.3 Link to CTSC Use-case: Grant & Pienaar at Children's Hospital Boston
- 1.4 Link to CTSC Use-case: Brad Dickerson at Mass General NMR
- 1.5 Link to CTSC Use-case: Warfield & Weisen at Children's
- 1.6 Link to CTSC Use-case: Managing Image Guided Therapy (NCIGT) Restrospective Data at Brigham & Women's Hospital
- 1.7 Link to FetchMI XNE extension planning (work with Curtis Lisle)
- 2 BIRN-related Presentations, Uses Cases and Pseudo Code
- 3 Development breadcrumbs and notes
- 4 Goal for how Slicer would upload/download from remote data stores =
- 5 Test-case URIs:
- 6 XNAT questions
 Link to Fetch Medical Informatics (FetchMI) implementation 1 notes
Link to Fetch Medical Informatics (FetchMI) implementation 1 notes
 Link to Fetch Medical Informatics (FetchMI) and RemoteIO profiling/refining notes
Link to Fetch Medical Informatics (FetchMI) and RemoteIO profiling/refining notes
 Link to CTSC Use-case: Grant & Pienaar at Children's Hospital Boston
Link to CTSC Use-case: Grant & Pienaar at Children's Hospital Boston
 Link to CTSC Use-case: Brad Dickerson at Mass General NMR
Link to CTSC Use-case: Brad Dickerson at Mass General NMR
 Link to CTSC Use-case: Warfield & Weisen at Children's
Link to CTSC Use-case: Warfield & Weisen at Children's
 Link to CTSC Use-case: Managing Image Guided Therapy (NCIGT) Restrospective Data at Brigham & Women's Hospital
Link to CTSC Use-case: Managing Image Guided Therapy (NCIGT) Restrospective Data at Brigham & Women's Hospital
 Link to FetchMI XNE extension planning (work with Curtis Lisle)
Link to FetchMI XNE extension planning (work with Curtis Lisle)
- March 2008 mBIRN meeting, XCEDE use cases
- March 2008 mBIRN meeting, pseudo code for web services
- April 2008 fBIRN meeting, Remote data handling architecture
- April 2008 fBIRN meeting, User-scenarios for web services and client tools discussion
Development breadcrumbs and notes
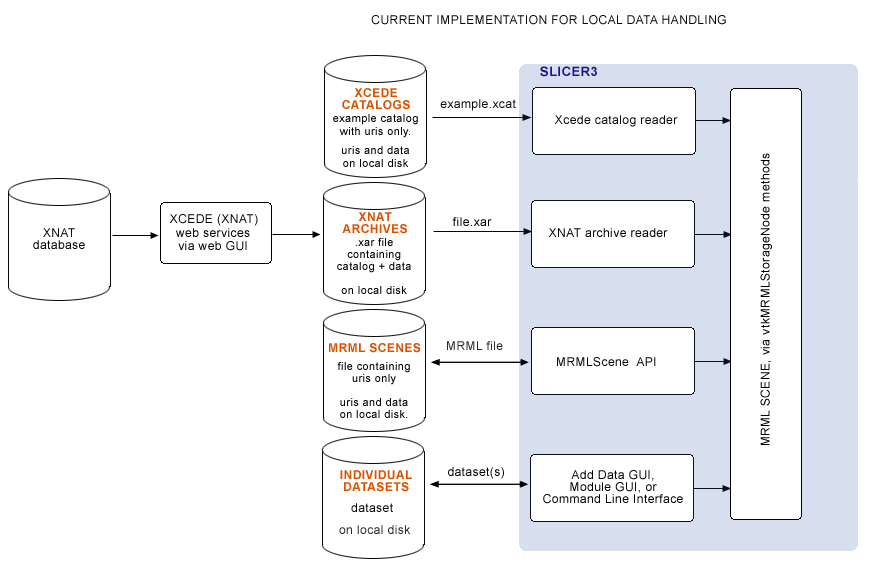
Slicer's original (local) data handling schematic
Originally, MRML files, XCEDE catalog files, XNAT archives and individual datasets were only loadable from local disk, and remote datasets were downloaded (via web interface or command line) outside of Slicer. In the BIRN 2007 AHM we demonstrated downloading .xar files from a remote database, and loading .xar and .xcat files into Slicer from local disk using Slicer's XNAT archive reader and XCEDE2.0 catalog reader. This original scheme for data handling is shown below:
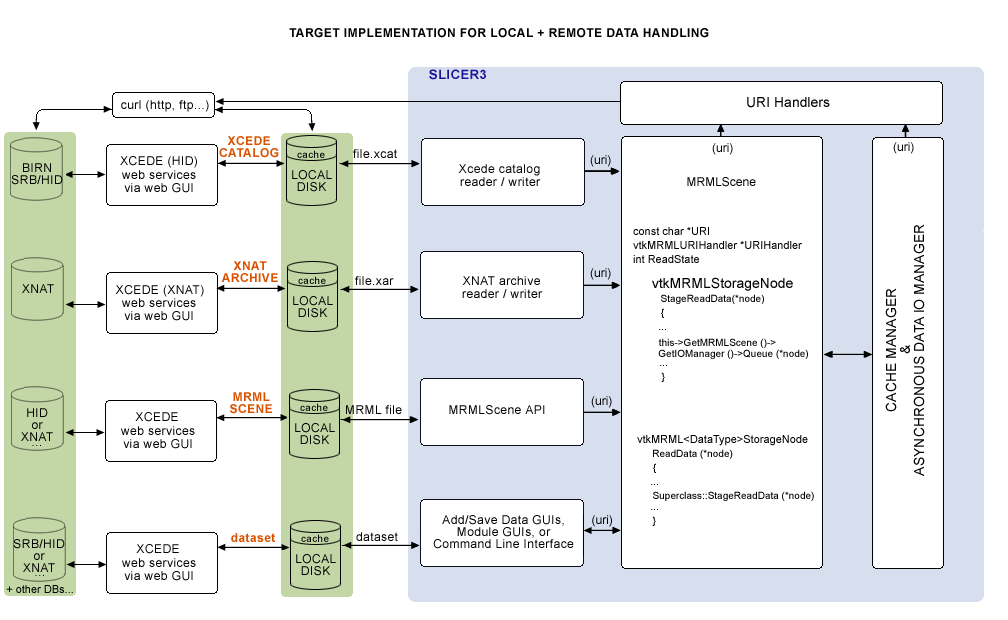
Goal for how Slicer would upload/download from remote data stores =
Eventually, we would like to query web services, download data remotely or locally from the Application itself, and have the option of uplaoding to remote stores as well. A sketch of the architecture planned in a meeting (on 2/14/08 with Steve Pieper, Nicole Aucoin and Wendy Plesniak) is shown below, including:
- a collection of vtkURIHandlers,
- an (asynchronous) Data I/O Manager and
- a Cache manager,
all created by the main application and pointed to by the MRMLScene.:
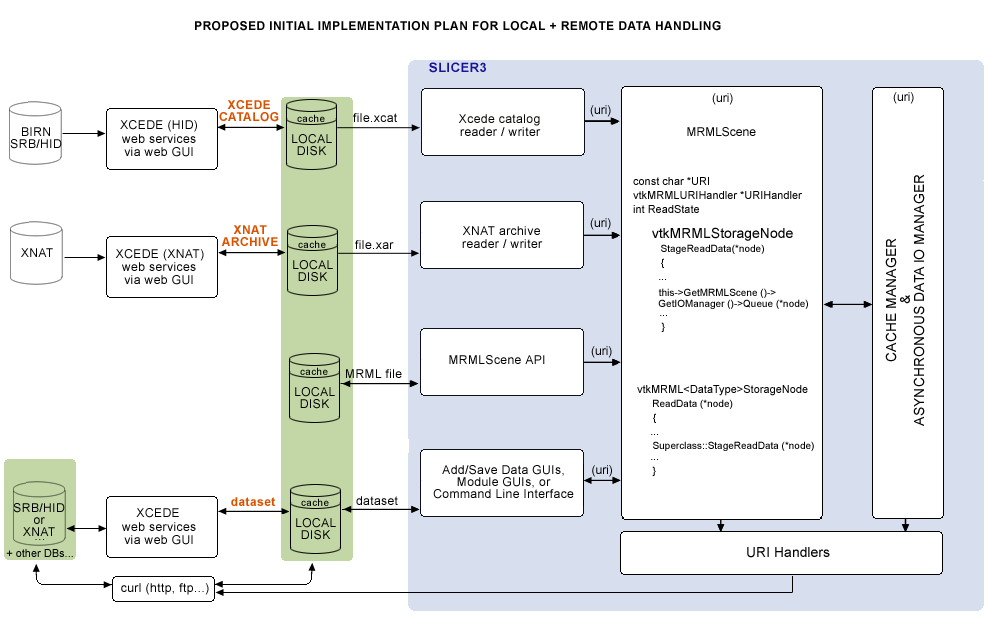
Two general use cases used to drive a first pass implementation
- First, is loading a combined FIPS/FreeSurfer analysis, specified in an Xcede catalog (.xcat) file that contains uris pointing to remote datasets, and view this with Slicer's QueryAtlas. (Prior to fBIRN AHM in 2008, it was not possible to get an .xcat via the HID web GUI; our approach was to manually upload a test Xcede catalog file and its constituent datasets to SRB. A copy of the .xcat file was kept locally, and SRB was accessed for each uri listed in it.)
- Second, is running a batch job in Slicer that processes a set of remotely held datasets. Each iteration would take as arguments the XML file parameterizing the EMSegmenter, the uri for the remote dataset, and a uri for storing back the segmentation results. This use case has not yet been implemented.
The schematic of the functionality we'll need is shown below:
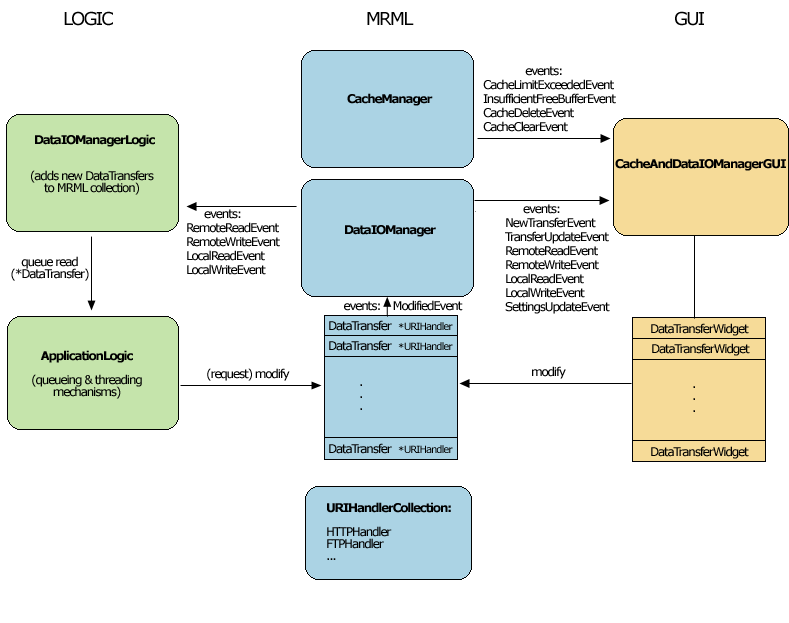
Current MRML, Logic and GUI Components
Slicer's Remote data handling architecture has been implemented to support remote download and upload of uris. Its components include a Cache Manager, a Data I/O Manager, and a URI Handler Collection in MRML; a Data I/O Manager Logic that engages the Application Logic's queueing and multi-threading mechanisms; and a Cache and Data I/O Manager GUI to provide feedback, information and control on the desktop. These components can also be configured through the Application Settings Interface, and these user preferences are saved in the Application Registry. Schematic and screenshots are shown below:
Test-case URIs:
- RemoteTest.mrml - points to CTHeadAxial.nrrd (the xml file is there, it may not render in your browser)
- RemoteTestVtk.mrml - points to Aseg_17_Left-Hippocampus.vtk, Aseg_53_Right-Hippocampus.vtk
- RemoteTestMgzVtk.mrml points to the above vtk files as well as Aseg.mgz
- RemoteTestColours.mrml points to Aseg.mgz and the colour file FSColorLabelsTest.ctbl
- RemoteTestMgzVtkPial.mrml points to Aseg.mgz, Aseg_53_Right-Hippocampus.vtk, and lh.pial
- RemoteTestSRBVtk.mrml points to a file on the BIRN SRB /home/naucoin.harvard-bwh/aseg_17_left-hippocampus.vtk (for now, requires that the uri string start with srb://) You must install and set up the SRB clients following the instructions here.
- RemoteBertSurfAndOverlays.mrml points to files on http://slicerl.bwh.harvard.edu/data, tests new overlay storage nodes'
- fBIRN-AHM2007.xcat a sample xcat file pointing to files on http://slicerl.bwh.harvard.edu/data/fBIRN (must download manually)
- BIRN-QA-XNAT.xcat a sample xcat file pointing to files on host http://central.xnat.org
- BIRN-QA-SRB.xcat points to files on the BIRN SRB /home/Public/FIPS-FreeSurfer-XCAT (for now, requires that the uri string start with srb://) You must install and set up the SRB clients following the instructions here.
- RemoteTwoFileVolumeSRB.mrml Points to a two part volume on the SRB (a test label map, should see three colours): /home/naucoin.harvard-bwh/segvolume.hdr and
/home/naucoin.harvard-bwh/segvolume.img
XNAT questions
- when we upload a file, can we get back a URI that includes ticket information? (we can use this to write a MRML file)
- can XNAT-generated uris end with the filename.extension, so that firefox will know to save them to disk, rather than to interpret them as html?
Implementation: new classes
MRML extensions
MRML-specific implementations and extensions of the following classes:
- vtkDataIOManager
- vtkMRMLStorageNode methods
- vtkMRML<DataType>StorageNode methods
- vtkPasswordPrompter
- vtkDataTransfer
- vtkCacheManager
- vtkURIHandler
GUI extensions
GUI implementations of the following classes:
- vtkSlicerDataTransferWidget
- vtkSlicerCacheAndDataIOManagerGUI
- vtkSlicerPasswordPrompter
Logic extensions
- vtkDataIOManagerLogic
Application Interface extensions
- Set CacheDirectory default = Slicer3 temp dir
- Set CacheFreeBufferSize default = 10Mb
- Set CacheLimit default = 20Mb
- Enable/Disable CacheOverwriting default = true
- Enable/Disable ForceRedownload default = false
- Enable/Disable asynchronous I/O default = false
- Instance & Register URI Handlers (?)
Important Notes
- MRML Scene file reading will always be synchronous.
- Read/write of individual datasets referenced in the scene should work with or without the asynchronous read/write turned on, and with or without the dataIOManager GUI interface.
ITK-based mechanism handling remote data (for command line modules, batch processing, and grid processing) (Nicole)
This one is tenatively on hold for now.
Workflows to support
The first goal is to figure out what workflows to support, and a good implementation approach.
Currently, Load Scene, Import Scene, and Add Data options in Slicer all encapsulate two steps:
- locating a dataset, usually accomplished through a file browser, and
- selecting a dataset to initiate loading.
Then MRML files, Xcede catalog files, or individual datasets are loaded from local disk.
For loading remote datasets, the following options are available:
- break these two steps apart explicitly (easiest option),
- bind them together under the hood,
- or support both of these paradigms.
For now, we choose the first option.
workflows:
Possible workflow A
- User downloads .xcat or .xml (MRML) file to disk using the HID or an XNAT web interface
- From the Load Scene file browser, user selects the .xcat or .xml archive. If no locally cached versions exist, each remote file listed in the archive is downloaded to /tmp directory (always locally cached) by the Data I/O Manager, and then cached (local) uri is passed to vtkMRMLStorageNode method when download is complete.
Possible workflow B
- User downloads .xcat or .xml (MRML) file to disk using the HID or an XNAT web interface
- From the Load Scene file browser, user selects the .xcat or .xml archive. If no locally cached versions exist, each remote file in the archive is downloaded to /tmp (only if a flag is set) by the Data IO Manager, and loaded directly into Slicer via a vtkMRMLStorageNode method when download is complete. (How does load work if we don't save to disk first?)
Workflow C
- describe batch processing example here, which includes saving to local or remote.
In each workflow, the data gets saved to disk first and then loaded into StorageNode or uploaded to remote location from cache.
What data do we need in an .xcat file?
For the fBIRN QueryAtlas use case, we need a combination of FreeSurfer morphology analysis and a FIPS analysis of the same subject. With the combined data in Slicer, we can view activation overlays co-registered to and overlayed onto the high resolution structural MRI using the FIPS analysis, and determine the names of brain regions where activations occur using the co-registered morphology analysis.
The required analyses including all derived data are in two standard directory structures on local disk, and *hopefully* somewhere on the HID within a standard structure (check with Burak). These directory trees contain a LOT of files we don't need... Below are the files we *do* need for fBIRN QueryAtlas use case.
FIPS analysis (.feat) directory and required data
For instance, the FIPS output directory in our example dataset from Doug Greve at MGH is called sirp-hp65-stc-to7-gam.feat. Under this directory, QueryAtlas needs the following datasets:
- sirp-hp65-stc-to7-gam.feat/reg/example_func.nii
- sirp-hp65-stc-to7-gam.feat/reg/freesurfer/anat2exf.register.dat
- sirp-hp65-stc-to7-gam.feat/stats/(all statistics files of interest)
- sirp-hp65-stc-to7-gam.feat/design.gif (this image relates statistics files to experimental conditions)
FreeSurfer analysis directory, and required data
For instance, the FreeSurfer morphology analysis directory in our example dataset from Doug Greve at MGH is called fbph2-000670986943. Under this directory, QueryAtlas needs the following datasets:
- fbph2-000670986943/mri/brain.mgz
- fbph2-000670986943/mri/aparc+aseg.mgz
- fbph2-000670986943/surf/lh.pial
- fbph2-000670986943/surf/rh.pial
- fbph2-000670986943/label/lh.aparc.annot
- fbph2-000670986943/label/rh.aparc.annot
What do we want HID webservices to provide?
- Question: are FIPS and FreeSurfer analyses (including QueryAtlas required files listed above) for subjects available on the HID yet? --Burak says not yet.
- Given that, can we manually upload an example .xcat and the datasets it points to the SRB, and download each dataset from the HID in Slicer, using some helper application (like curl)?
- (Eventually.) The BIRN HID webservices shouldn't really need to know the subset of data that QueryAtlas needs... maybe the web interface can take a BIRN ID and create a FIPS/FreeSurfer xcede catalog with all uris (http://....) in the FIPS and FreeSurfer directories, and package these into an Xcede catalog.
- (Eventually.) The catalog could be requested and downloaded from the HID web GUI, with a name like .xcat or .xcat.gzip or whatever. QueryAtlas could then open this file (or unzip and open) and filter for the relevant uris for an fBIRN or Qdec QueryAtlas session.
- Then, for each uri in a catalog (or .xml MRML file), we'll use (curl?) to download; so we need all datasets to be publicly readable.
- Can we create a directory (even a temporary one) on the SRB/BWH HID for Slicer data uploads?
- We need some kind of upload service, a function call that takes a dataset and a BIRNID and uploads data to appropriate remote directory.