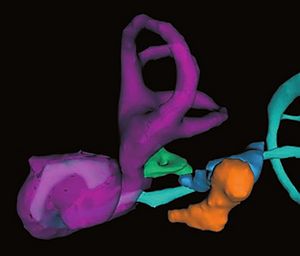Special:Badtitle/NS100:Slicer Community
Contents
- 1 3D Slicer Enabled Research
- 2 2013
- 2.1 Fiber Tract-Driven Topographical Mapping (Fttm) Reveals Microstructural Relevance for Interhemispheric Visuomotor Function In the Aging Brain
- 2.2 Prostate Volumetric Assessment by Magnetic Resonance Imaging and Transrectal Ultrasound: Impact of Variation in Calculated Prostate-Specific Antigen Density on Patient Eligibility for Active Surveillance Program
- 2.3 Optic Radiation Fiber Tractography in Glioma Patients Based on High Angular Resolution Diffusion Imaging with Compressed Sensing Compared with Diffusion Tensor Imaging - Initial Experience
- 2.4 Augmented Reality Visualization Using Image Overlay Technology for MR-Guided Interventions: Cadaveric Bone Biopsy at 1.5 T
- 2.5 Clinical correlates of nucleus accumbens volume in drug-naive, adult patients with obsessive-compulsive disorder
- 2.6 Retrosigmoid implantation of an active bone conduction stimulator in a patient with chronic otitis media
- 2.7 Computed tomography image processing to detect the real mechanism of bioprosthesis failure: implication for valve-in-valve implantation
- 2.8 Preliminary Study on the Clinical Application of Augmented Reality Neuronavigation
- 2.9 Can transrectal needle biopsy be optimised to detect nearly all prostate cancer with a volume of ≥0.5 mL? A three-dimensional analysis
- 2.10 Quantitative MRI Analysis of Craniofacial Bone Marrow in Patients with Sickle Cell Disease
- 2.11 Three-dimensional volumetric rendition of cannon ball pulmonary metastases by the use of 3D Slicer, an open source free software package
- 2.12 MRI Evidence for Altered Venous Drainage and Intracranial Compliance in Mild Traumatic Brain Injury
- 2.13 Augmented reality visualisation using an image overlay system for MR-guided interventions: technical performance of spine injection procedures in human cadavers at 1.5 Tesla
- 3 2012
- 3.1 Perk Tutor: an open-source training platform for ultrasound-guided needle insertions
- 3.2 Pituitary Adenoma Volumetry with 3D Slicer
- 3.3 Differences in simple morphological variables in ruptured and unruptured middle cerebral artery aneurysms
- 3.4 3D Slicer as an image computing platform for the Quantitative Imaging Network
- 3.5 SlicerRT: radiation therapy research toolkit for 3D Slicer
- 3.6 Developmental Trajectories of Amygdala and Hippocampus from Infancy to Early Adulthood in Healthy Individuals
- 3.7 Extra-Hippocampal Subcortical Limbic Involvement Predicts Episodic Recall Performance in Multiple Sclerosis
- 3.8 Posterior vaginal prolapse shape and position changes at maximal Valsalva seen in 3-D MRI-based models.
- 3.9 Talairach methodology in the multimodal imaging and robotics era
- 3.10 SARRP Dose Planning System
- 3.11 Three-dimensional image quantification as a new morphometry method for microvascular tissue engineering
- 3.12 Prostate: registration of digital histopathologic images to in vivo MR images acquired by using endorectal receive coil
- 3.13 A comparison of the effect of age on levator ani and obturator internus muscle cross-sectional areas and volumes in nulliparous women
- 3.14 Levator ani subtended volume: a novel parameter to evaluate levator ani muscle laxity in pelvic organ prolapse
- 3.15 Tractography Delineates Microstructural Changes in the Trigeminal Nerve after Focal Radiosurgery for Trigeminal Neuralgia
- 3.16 Square-cut: A Segmentation Algorithm on the Basis of a Rectangle Shape
- 3.17 Anatomical and functional correlates of human hippocampal volume asymmetry
- 3.18 Hippocampal subregions are differentially affected in the progression to Alzheimer's disease
- 3.19 Long-term memory search across the visual brain
- 3.20 Temporal course of cerebrospinal fluid dynamics and amyloid accumulation in the aging rat brain from three to thirty months
- 3.21 Intact relational memory and normal hippocampal structure in the early stage of psychosis
- 3.22 TREK: an integrated system architecture for intraoperative cone-beam CT-guided surgery
- 4 2011
- 4.1 Atlas-guided segmentation of vervet monkey brain MRI
- 4.2 A System for Video-based Navigation for Endoscopic Endonasal Skull Base Surgery
- 4.3 Comparison of Acute and Chronic Traumatic Brain Injury using Semi-automatic Multimodal Segmentation of MR Volumes
- 4.4 3D Slicer
- 4.5 An Open Environment CT-US Fusion for Tissue Segmentation during Interventional Guidance
- 4.6 Mixed handedness is associated with greater age-related decline in volumes of the hippocampus and amygdala: the PATH through life study
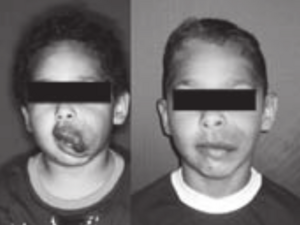
- 4.7 Multiple Costal Cartilage Graft Reconstruction for the Treatment of a Full-length Laryngotracheal Stenosis after an Inhalation Burn
- 4.8 A supervised patch-based approach for human brain labeling
- 4.9 The chronic bronchitic phenotype of COPD: an analysis of the COPDGene Study
- 4.10 Airway Inspector: Phenotyping the Lung in COPD
- 4.11 Distance Measurement in Middle Ear Surgery using a Telemanipulator
- 4.12 3D Slicer as a tool for interactive brain tumor segmentation
- 4.13 The Effect of Augmented Reality Training on Percutaneous Needle Placement in Spinal Facet Joint Injections
- 4.14 Optimal transseptal puncture location for robot-assisted left atrial catheter ablation
- 4.15 Multiple indices of diffusion identifies white matter damage in mild cognitive impairment and Alzheimer's disease
- 4.16 Assessment of abdominal adipose tissue and organ fat content by magnetic resonance imaging
- 4.17 Consistent neuroanatomical age-related volume differences across multiple samples
- 4.18 Brain Regional Lesion Burden and Impaired Mobility in the Elderly
- 4.19 Lung Volumes and Emphysema in Smokers with Interstitial Lung Abnormalities
- 4.20 Integration of 3D anatomical data obtained by CT imaging and 3D optical scanning for computer aided implant surgery
- 4.21 Anisotropy of Transcallosal Motor Fibres Indicates Functional Impairment in Children with Periventricular Leukomalacia
- 4.22 Automatic lung segmentation in CT images with accurate handling of the hilar region
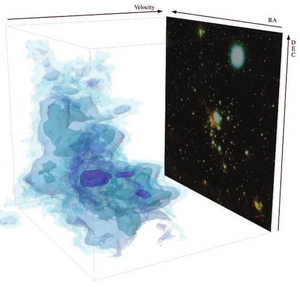
- 4.23 Using 3D Slicer in Astronomy
- 4.24 Brain morphometry in autism spectrum disorders: a unified approach for structure-specific statistical analysis of neuroimaging data
- 4.25 Three dimensional in vivo modelling of vestibular schwannomas and surrounding cranial nerves using diffusion imaging tractography
- 4.26 Intraoperative Real-Time Querying of White Matter Tracts During Frameless Stereotactic Neuronavigation
- 5 2010
- 5.1 Application of Vascular Model Toolkit (VMTK) for Coronary Arteries
- 5.2 Ion-Abrasion Scanning Electron Microscopy Reveals Surface-Connected Tubular Conduits in HIV-Infected Macrophages
- 5.3 MRI-Guided Robotic Prostate Biopsy: A Clinical Accuracy Validation
- 5.4 A method for planning safe trajectories in image-guided keyhole neurosurgery
- 5.5 JHU Workshop for Talented Youth
- 5.6 Canine Hippocampal Formation Composited into Three-dimensional Structure Using MPRAGE
- 5.7 Real-time 3-dimensional virtual reality navigation system with open MRI for breast-conserving surgery
- 5.8 Interfaces and Integration of Medical Image Analysis Frameworks: Challenges and Opportunities
- 5.9 Evaluation of Robotic Needle Steering in ex vivo Tissue
- 5.10 An open source implementation of colon CAD in 3D Slicer
- 5.11 In vivo visualization of cranial nerve pathways in humans using diffusion-based tractography
- 5.12 Coronary Artery Centerline Extraction in 3D Slicer using VMTK based Tools
- 5.13 Assessment of Image Registration Accuracy in Three-dimensional Transrectal Ultrasound Guided Prostate Biopsy
- 5.14 Language laterality in autism spectrum disorder and typical controls: a functional, volumetric, and diffusion tensor MRI study
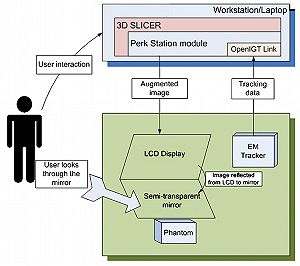
- 5.15 Perk Station-Percutaneous Surgery Training and Performance Measurement Platform
- 6 2009
- 6.1 Three-dimensional appearance of the lips muscles with three-dimensional isotropic MRI: in vivo study
- 6.2 Traces
- 6.3 CLIMB study (Comprehensive Longitudinal Investigation of Multiple Sclerosis at Brigham and Women’s Hospital) former NHS study (The Harvard Multiple Sclerosis Natural History Study)
- 6.4 Steroid Adjunctive Treatment at Initiation of Avonex Therapy for Patients with Mono-Symptomatic or Relapsing-Remitting Multiple Sclerosis
- 6.5 Age-Related Changes of Cognition in Health and Diseases: Image Analysis Core
- 6.6 Harvard Research Nursing Home Project (MOBILIZE study)
- 6.7 Daclizumab Use in Patients with Pediatric Multiple Sclerosis Failing Interferon
- 6.8 MRI findings of pediatric-onset Multiple Sclerosis patients: a retrospective study
- 6.9 Pediatric MS MRI Pilot Project
- 6.10 The Multiple Sclerosis Database Project (http://cni-boston.org)
- 6.11 Effects of Strain Thresholds on Bone Formation in Response to Mechanical Loading
- 6.12 Automated ventricular systems segmentation in brain CT images by combining low-level segmentation and high-level template matching
- 6.13 MRI-derived measurements of human subcortical, ventricular and intracranial brain volumes: Reliability effects of scan sessions, acquisition sequences, data analyses, scanner upgrade, scanner vendors and field strengths
- 6.14 Integration of the Vascular Modeling Toolkit in 3D Slicer
- 6.15 New EMSegment Module in Slicer3
- 6.16 Interactive 3D Navigation System for Image-guided Surgery
- 6.17 Smaller Amygdala is Associated with Anxiety in Patients with Panic Disorder
- 6.18 Optimal Transseptal Puncture Location for Robot-Assisted Left Atrial Catheter Ablation
- 6.19 Measurements from image-based three dimensional pelvic floor reconstruction: a study of inter- and intraobserver reliability
- 6.20 Three-dimensional analysis of rodent paranasal sinus cavities from X-ray computed tomography (CT) scans
- 6.21 Multimodal imaging in mild cognitive impairment: Metabolism, morphometry and diffusion of the temporal–parietal memory network
- 6.22 The relation between connection length and degree of connectivity in young adults: a DTI analysis
- 6.23 The ROBOCAST project: ROBOt and sensors integration for Computer Assisted Surgery and Therapy
- 6.24 A trajectory planning method for reduced patient risk in image-guided neurosurgery: preliminary results
- 6.25 Contrast-maximizing adaptive region growing for CT
- 6.26 Computer-aided 3D visualization in oto-rhino-laryngology
- 6.27 Clinical Application of Curvilinear Distraction Osteogenesis for Correction of Mandibular Deformities
- 6.28 In Vivo Hippocampal Measurement and Memory: A Comparison of Manual Tracing and Automated Segmentation in a Large Community-Based Sample
- 6.29 How volumetric analysis quantifies therapeutic response of slow-flow vascular malformations
- 6.30 A minimally invasive registration method using surface template-assisted marker positioning (STAMP) for image-guided otologic surgery
- 6.31 Diffusion Tractography of the Fornix in Schizophrenia
- 6.32 Spiny Versus Stubby: 3D Reconstruction of Human Myenteric (type I) Neurons
- 7 2008
- 7.1 An Integrated System for Planning, Navigation and Robotic Assistance for Skull Base Surgery
- 7.2 The relationship between diffusion tensor imaging and volumetry as measures of white matter properties
- 7.3 3D modeling-based surgical planning in transsphenoidal pituitary surgery--preliminary results
- 7.4 A Generic Framework for Internet-based Interactive Applications of High-resolution 3-D Medical Image Data
- 7.5 Anterior cingulate cortex volume reduction in patients with panic disorder
- 7.6 A computer modelling tool for comparing novel ICD electrode orientations in children and adults
- 7.7 Using magnetic resonance microscopy to study the growth dynamics of a glioma spheroid in collagen I: A case study
- 7.8 Lowering the Barriers Inherent in Translating Advances in Neuroimage Analysis to Clinical Research Applications
- 8 2007
- 8.1 Towards scarless surgery: An endoscopic ultrasound navigation system for transgastric access procedures
- 8.2 3D visualization and simulation of frontoorbital advancement in metopic synostosis
- 8.3 Creating physical 3D stereolithograph models of brain and skull]
- 8.4 Morphology, constraints, and scaling of frontal sinuses in the hartebeest, Alcelaphus buselaphus (Mammalia: Artiodactlya, Bovidae)
- 8.5 Image-guided Otologic Surgery based on Patient Motion Compensation and Intraoperative Virtual CT
- 8.6 Non-rigid registration of pre-procedural MR images with intra- procedural unenhanced CT images for improved targeting of tumors during liver radiofrequency ablations
- 8.7 A detailed 3D model of the guinea pig cochlea
- 8.8 Dynamic 3-D computer modeling tracks brain changes during surgery
- 8.9 Surface Rendering-based Virtual Intraventricular Endoscopy: Retrospective Feasibility Study and Comparison to Volume Rendering-based Approach
- 8.10 Interest of the Steady State Free Precession (SSFP) sequence for 3D modeling of the whole fetus
- 8.11 Longitudinal in vivo Reproducibility of Cartilage Volume and Surface in Osteoarthritis of the Knee
- 8.12 Origin and insertion points involved in levator ani muscle defects
- 8.13 A prototype biosensor-integrated image-guided surgery system
- 8.14 Accuracy evaluation of initialization-free registration for intraoperative 3D-navigation
- 8.15 Development of a three-dimensional multiscale agent-based tumor model: simulating gene-protein interaction profiles, cell phenotypes and multicellular patterns in brain cancer
- 8.16 Image Registered FAST (IRFAST) for Combat Casualty Triage
- 8.17 A Preliminary Study on the Relationship between Nasal Cavity and Maxillary Sinus Volumes
- 8.18 Interobserver variability in the determination of upper lobe-predominant emphysema
- 8.19 Registered, Sensor-Integrated Virtual Reality for Surgical Applications
- 8.20 Modeling Cancer Biology
- 8.21 A Comparison of Upper Airway Structures in Male and Female Obstructive Sleep Apnoea (OSA) Patients
- 9 2006
- 9.1 Shaving diffusion tensor images in discriminant analysis: a study into schizophrenia
- 9.2 Interventional navigation for abdominal surgery by simultaneous use of MRI and ultrasound
- 9.3 Molar crown thickness, volume, and development in South African Middle Stone Age humans
- 9.4 A 3D Model Simulating Sediment Transport, Erosion and Deposition within a Network of Channel Belts and an Associated Floodplain
- 9.5 Molecular Diffusion in MRI: Technical Application of Fiber Tracking
- 9.6 Quantification of Levator Ani Cross-sectional Area Differences between Women with and those without Prolapse
- 9.7 Liver metastases: 3D shape-based analysis of CT scans for detection of local recurrence after radiofrequency ablation
- 9.8 Atlas Guided Identification of Brain Structures by Combining 3D Segmentation and SVM Classification
- 9.9 Development of a CAD (Computer Assisted Detection) System to Detect Lung Nodules in CT Scans
- 9.10 Dynamic Simulation of Joints Using Multi-Scale Modeling
- 9.11 Quasi-isometric Flattening of Curved Surfaces for Medical Imaging
- 9.12 A Translation Station for Imaging
- 9.13 User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability.
- 9.14 Habitual Use of the Primate Forelimb is Reflected in the Material Properties of Subchondral Bone in the Distal Radius
- 9.15 Appearance of the Levator Ani Muscle Subdivisions in Magnetic Resonance Images
- 9.16 Virtual Cystoscopy - A Surgical Planning and Guidance Tool
- 9.17 Measurement of the Pubic Portion of the Levator ani Muscle in Women with Unilateral Defects in 3D Models from MR Images
- 9.18 Finite-element-method (FEM) Model Generation of Time-resolved 3D Echocardiographic Geometry Data for Mitral-valve Volumetry
- 9.19 Range of Curvilinear Distraction Devices Required for Treatment of Mandibular Deformities
- 9.20 Preliminary Study on Digitized Nasal and Temporal Bone Anatomy
- 9.21 Developmental Response to Cold Stress in Cranial Morphology of Rattus: Implications for the Interpretation of Climatic Adaptation in Fossil Hominins
- 9.22 A ceratopsid dinosaur parietal from New Mexico and its implications for ceratopsid biogeography and systematics
- 10 2005
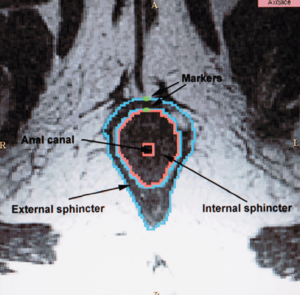
- 10.1 Magnetic Resonance Imaging and 3-Dimensional Analysis of External Anal Sphincter Anatomy
- 10.2 Group-Slicer: a collaborative extension of 3D Slicer
- 10.3 2D Rigid Registration of MR Scans using the 1D Binary Projections
- 10.4 Three-dimensional reconstruction and volumetry of intracranial haemorrhage and its mass effect
- 10.5 Vaginal Thickness, Cross-Sectional Area, and Perimeter in Women with and Those without Prolapse
- 10.6 Registration and Fusion of CT and MRI of the Temporal Bone
- 10.7 Open-configuration MR-guided Microwave Thermocoagulation Therapy for Metastatic Liver Tumors from Breast Cancer
- 10.8 Quantification of Airway Diameters and 3D Airway Tree Rendering from Dynamic Hyperpolarized 3He Magnetic Resonance Imaging
- 10.9 A finite element method model to simulate laser interstitial thermo therapy in anatomical inhomogeneous regions
- 10.10 The Application of DTI to Investigate White Matter Abnormalities in Schizophrenia
- 11 2004
- 11.1 A Statistically Based Flow for Image Segmentation
- 11.2 Decreases in Ventricular Volume Correlate with Decreases in Ventricular Pressure in Idiopathic Normal Pressure Hydrocephalus Patients who Experienced Clinical Improvement after Implantation with Adjustable Valve Shunts
- 11.3 Spatial Motion Constraints in Medical Robot Using Virtual Fixtures Generated by Anatomy
- 11.4 Clinical validation of the normalized mutual information method for registration of CT and MR images in radiotherapy of brain tumors.
- 11.5 Diffusion-tensor imaging–guided tracking of fibers of the pyramidal tract combined with intraoperative cortical stimulation mapping in patients with gliomas
- 11.6 Abnormal Association between Reduced Magnetic Mismatch Field to Speech Sounds and Smaller Left Planum Temporale Volume in Schizophrenia
- 12 2003
- 12.1 Genetic contribution to cartilage volume in women: a classical twin study
- 12.2 Advanced computer assistance for magnetic resonance-guided microwave thermocoagulation of liver tumors
- 12.3 The association of cartilage volume with knee pain
- 12.4 Three-dimensional reconstruction of magnetic resonance images of the anal sphincter and correlation between sphincter volume and pressure
- 12.5 Surgical Navigation in the Open MRI
- 12.6 The virtual craniofacial patient: 3D jaw modeling and animation
- 13 2002
- 14 2001
- 15 2000
3D Slicer Enabled Research
3D Slicer is a free open source software package distributed under a BSD style license. The majority of funding for the development of 3D slicer comes from a number of grants and contracts from the National Institutes of Health. See Slicer Acknowledgments for more information.
This page focuses on research that was done outside of our immediate collaboration community. That community is represented in the publication database.
We invite you to provide information on how you are using 3D Slicer to produce peer-reviewed research. Information about the scientific impact of this tool is helpful in raising funding for the continued support of this tool.
2013
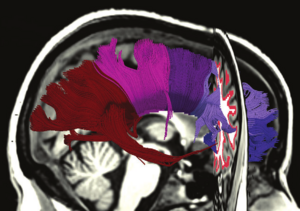
Fiber Tract-Driven Topographical Mapping (Fttm) Reveals Microstructural Relevance for Interhemispheric Visuomotor Function In the Aging Brain
|
Publication: Neuroimage. 2013 Aug 15;77:195-206. PMID: 23567886 Authors: Schulte T, Maddah M, Müller-Oehring EM, Rohlfing T, Pfefferbaum A, Sullivan EV. Institution: SRI International, Neuroscience Program, Menlo Park, CA, USA. Background/Purpose: We present a novel approach - DTI-based fiber tract-driven topographical mapping (FTTM) - to map and measure the influence of age on the integrity of interhemispheric fibers and challenge their selective functions with measures of interhemispheric integration of lateralized information. This approach enabled identification of spatially specific topographical maps of scalar diffusion measures and their relation to measures of visuomotor performance. Relative to younger adults, older adults showed lower fiber integrity indices in anterior than posterior callosal fibers. FTTM analysis identified a dissociation in the microstructural-function associates between age groups: in younger adults, genu fiber integrity correlated with interhemispheric transfer time, whereas in older adults, body fiber integrity was correlated with interhemispheric transfer time with topographical specificity along left-lateralized callosal fiber trajectories. Neural co-activation from redundant targets was evidenced by fMRI-derived bilateral extrastriate cortex activation in both groups, and a group difference emerged for a pontine activation cluster that was differently modulated by response hand in older than younger adults. Bilateral processing advantages in older but not younger adults further correlated with fiber integrity in transverse pontine fibers that branch into the right cerebellar cortex, thereby supporting a role for the pons in interhemispheric facilitation. In conclusion, in the face of compromised anterior callosal fibers, older adults appear to use alternative pathways to accomplish visuomotor interhemispheric information transfer and integration for lateralized processing. This shift from youthful associations may indicate recruitment of compensatory mechanisms involving medial corpus callosum fibers and subcortical pathways. Fundings:
|
Prostate Volumetric Assessment by Magnetic Resonance Imaging and Transrectal Ultrasound: Impact of Variation in Calculated Prostate-Specific Antigen Density on Patient Eligibility for Active Surveillance Program
|
Publication: Journal of Computer Assisted Tomography. 2013 July/August;37(4):589-595. Authors: Dianat, Seyed Saeid, Rancier Ruiz, Ramiro M., Bonekamp, David, Carter, H. Ballentine, Macura, Katarzyna J. Institution: Department of Radiology, Johns Hopkins University, Baltimore, MD. Background/Purpose: Objective: The objective of this study was to investigate impact of prostate volume variations on prostate-specific antigen density (PSAD) and patient eligibility for active surveillance (AS). Methods: Prostate volume and PSAD were calculated for 46 patients with prostate cancer in AS who underwent prostate magnetic resonance imaging and transrectal ultrasound (TRUS). Manual method and 2 semiautomated methods for prostate segmentation (3D Slicer and OsiriX) were used for MR volumetry. Results: Magnetic resonance volumetric methods showed very good agreement (intraclass correlation coefficient, 0.98). The concordance correlation coefficient was higher among MR volumetry methods (0.971–0.998) than between TRUS and MR volumetry (0.849–0.863). The variation in PSAD estimated by TRUS versus magnetic resonance imaging was higher in large prostates (r = 0.327, P = 0.027). Transrectal ultrasonography volumetry may improperly classify 20% of patients as eligible for AS with PSAD greater than 0.15 threshold. Conclusions: Although clinically used TRUS reliably estimates PSAD, it may misclassify some patients who are not eligible for AS based on PSAD criteria. Magnetic resonance–based volumetry should be considered for a more reliable PSAD calculation. |
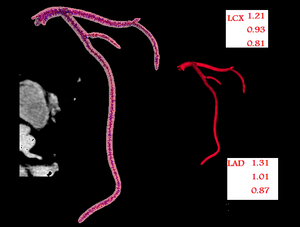
Optic Radiation Fiber Tractography in Glioma Patients Based on High Angular Resolution Diffusion Imaging with Compressed Sensing Compared with Diffusion Tensor Imaging - Initial Experience
|
Publication: PLoS One. 2013 July;8(7):e70973. | PDF Authors: Kuhnt D, Bauer M, Sommer J, Merhof D, Nimsky C. Institution: Department of Neurosurgery, University of Marburg, Marburg, Germany. Background/Purpose: Up to now, fiber tractography in the clinical routine is mostly based on diffusion tensor imaging (DTI). However, there are known drawbacks in the resolution of crossing or kissing fibers and in the vicinity of a tumor or edema. These restrictions can be overcome by tractography based on High Angular Resolution Diffusion Imaging (HARDI) which in turn requires larger numbers of gradients resulting in longer acquisition times. Using compressed sensing (CS) techniques, HARDI signals can be obtained by using less non-collinear diffusion gradients, thus enabling the use of HARDI-based fiber tractography in the clinical routine. Eight patients with gliomas in the temporal lobe, in proximity to the optic radiation (OR), underwent 3T MRI including a diffusion-weighted dataset with 30 gradient directions. Fiber tractography of the OR using a deterministic streamline algorithm based on DTI was compared to tractography based on reconstructed diffusion signals using HARDI+CS. HARDI+CS based tractography displayed the OR more conclusively compared to the DTI-based results in all eight cases. In particular, the potential of HARDI+CS-based tractography was observed for cases of high grade gliomas with significant peritumoral edema, larger tumor size or closer proximity of tumor and reconstructed fiber tract. Overcoming the problem of long acquisition times, HARDI+CS seems to be a promising basis for fiber tractography of the OR in regions of disturbed diffusion, areas of high interest in glioma surgery. Funding:
|
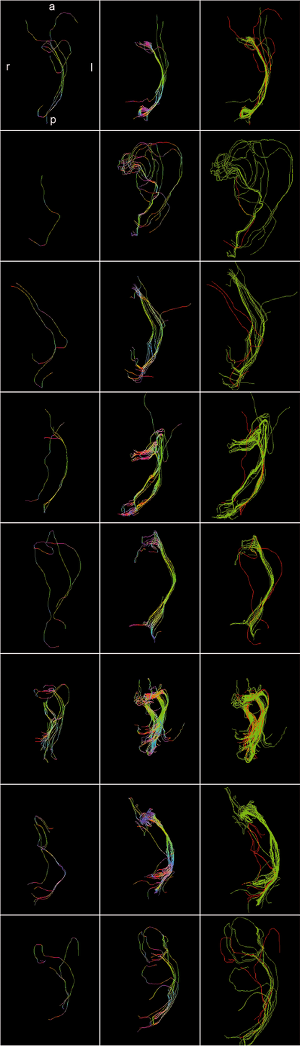 Fiber tractography results presented for each patient (patients 1–8 according rows 1–8) based on DTI (column 1), Slicer 4 (column 2), and based on HARDI+CS (column 4) within MedAlyVis. Overlay of DTI-based (red) and HARDI+CS-based tractography (green) (r = right; l = left; a = anterior; p = posterior). |
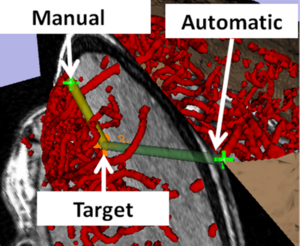
Augmented Reality Visualization Using Image Overlay Technology for MR-Guided Interventions: Cadaveric Bone Biopsy at 1.5 T
|
Publication: Invest Radiol. 2013 Jun;48(6):464-470. PMID: 23328911 Authors: Fritz J, U-Thainual P, Ungi T, Flammang AJ, McCarthy EF, Fichtinger G, Iordachita II, Carrino JA. Institution: Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, MD, USA. Background/Purpose: The purpose of this study was to prospectively test the hypothesis that image overlay technology facilitates accurate navigation for magnetic resonance (MR)-guided osseous biopsy. A prototype augmented reality image overlay system was used in conjunction with a clinical 1.5-T MR imaging system. Osseous biopsy of a total of 16 lesions was planned in 4 human cadavers with osseous metastases. A loadable module of 3D Slicer open-source medical image analysis and visualization software was developed and used for display of MR images, lesion identification, planning of virtual biopsy paths, and navigation of drill placement. The osseous drill biopsy was performed by maneuvering the drill along the displayed MR image containing the virtual biopsy path into the target. The drill placement and the final drill position were monitored by intermittent MR imaging. Outcome variables included successful drill placement, number of intermittent MR imaging control steps, target error, number of performed passes and tissue sampling, time requirements, and pathological analysis of the obtained osseous core specimens including adequacy of specimens, presence of tumor cells, and degree of necrosis. A total of 16 osseous lesions were sampled with percutaneous osseous drill biopsy. Eight lesions were located in the osseous pelvis (8/16, 50%) and 8 (8/16, 50%) lesions were located in the thoracic and lumbar spine. Lesion size was 2.2 cm (1.1-3.5 cm). Four (2-8) MR imaging control steps were required. MR imaging demonstrated successful drill placement inside 16 of the 16 target lesions (100%). One needle pass was sufficient for accurate targeting of all lesions. One tissue sample was obtained in 8 of the 16 lesions (50%); 2, in 6 of the16 lesions (38%); and 3, in 2 of the 16 lesions (12%). The target error was 4.3 mm (0.8-6.8 mm). Length of time required for biopsy of a single lesion was 38 minutes (20-55 minutes). Specimens of 15 of the 16 lesions (94%) were sufficient for pathological evaluation. Of those 15 diagnostic specimens, 14 (93%) contained neoplastic cells, whereas 1 (7%) specimen demonstrated bone marrow without evidence of neoplastic cells. Of those 14 diagnostic specimens, 11 (79%) were diagnostic for carcinoma or adenocarcinoma, which was concordant with the primary neoplasm, whereas, in 3 of the 14 diagnostic specimens (21%), the neoplastic cells were indeterminate. Image overlay technology provided accurate navigation for the MR-guided biopsy of osseous lesions of the spine and the pelvis in human cadavers at 1.5 T. The high technical and diagnostic yield supports further evaluation with clinical trials. |
Clinical correlates of nucleus accumbens volume in drug-naive, adult patients with obsessive-compulsive disorder
|
Publication: Aust N Z J Psychiatry. 2013 Jun 4. PMID: 23737599 Authors: Narayanaswamy JC, Jose D, Kalmady S, Venkatasubramanian G, Reddy YJ. Institution: Obsessive Compulsive Disorder Clinic & Translational Psychiatry Lab, Neurobiology Research Center at National Institute of Mental Health and Neurosciences (NIMHANS), Bangalore, India. Background/Purpose: Background:Reward-processing deficits have been demonstrated in obsessive-compulsive disorder (OCD) and this has been linked to ventral striatal abnormalities. However, volumetric abnormalities of the nucleus accumbens (NAcc), a key structure in the reward pathway, have not been examined in OCD. We report on the volumetric abnormalities of NAcc and its correlation with illness severity in drug-naïve, adult patients with OCD.Method:In this cross-sectional study of case-control design, the magnetic resonance imaging (MRI) 1.5-T (1-mm) volume of NAcc was measured using 3D Slicer software in drug-naïve OCD patients (n = 44) and age, sex and handedness-matched healthy controls (HCs) (n = 36) using a valid and reliable method. OCD symptoms were assessed using the Yale-Brown Obsessive Compulsive Scale (Y-BOCS) Symptom checklist and severity and the Clinical Global Impression-Severity (CGI-S) scale.Results:There was no significant difference in NAcc volumes on either side between OCD patients and HCs (F = 3.45, p = 0.07). However, there was significant negative correlation between the right NAcc volume and Y-BOCS compulsion score (r = -0.48, p = 0.001).Conclusions:Study observations suggest involvement of the NAcc in the pathogenesis of OCD, indicating potential reward-processing deficits. Correlation between the right NAcc volume deficit and severity of compulsions offers further support for this region as a candidate for deep brain stimulation treatment in OCD. |
Retrosigmoid implantation of an active bone conduction stimulator in a patient with chronic otitis media
|
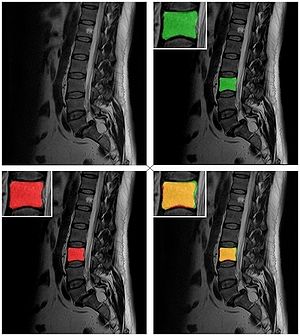
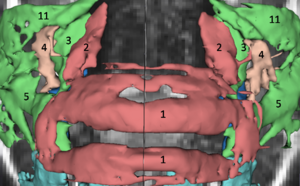
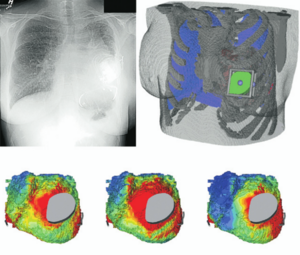
Publication: Auris Nasus Larynx. 2013 May 27. PMID: 23722197 Authors: Lassaletta L, Sanchez-Cuadrado I, Muñoz E, Gavilan J. Institution: Department of Otolaryngology, "La Paz" University Hospital, IdiPaz Research Institute, Madrid, Spain. Background/Purpose: Percutaneous bone conduction implants are widely used in patients with conductive and mixed hearing loss with no benefit from conventional air conduction hearing aids. These devices have several complications including skin reaction, wound infection, growth of skin over the abutment, and implant extrusion. We describe a case of a transcutaneous bone conduction implantation (Bonebridge, Med-el) in a patient with conductive hearing loss due to chronic otitis media. Surgical planification was performed with the software 3D slicer 4.1. According to this program, the implant transductor was positioned in the retrosigmoid area. Aided thresholds demonstrate a significant benefit, with an improvement from 68dB to 25dB. Speech discrimination scores improved 35dB. The patient is very happy and uses her device daily. The Bonebridge implant is a promising transcutaneous bone conduction implant for patients with conductive hearing loss. Retrosigmoid implantation may be useful in cases with mastoid pathology or previous surgery. |
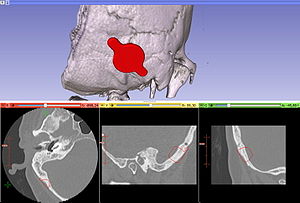 Top: 3D slicer reconstruction showing the planned location of the transductor (red) in the right retrosigmoid area. The theoretical position and area of dura compression (red figure) is shown in the axial (bottom, left), coronal (bottom, middle) and sagital (bottom, right) CT scan images. Note the planned position of one of the screws in the coronal view. |
Computed tomography image processing to detect the real mechanism of bioprosthesis failure: implication for valve-in-valve implantation
|
Publication: J Heart Valve Dis. 2013 Mar;22(2):236-8. PMID: 23798214 Authors: Ruggieri VG, Anselmi A, Wang Q, Esneault S, Haigron P, Verhoye JP. Institution: Department of Cardiovascular and Thoracic Surgery, Pontchaillou University Hospital, Rennes, France. Background/Purpose: Transcatheter valve-in-valve implantation is an emerging option for patients with structural deterioration of an aortic bioprosthesis and who are at a high surgical risk. The present case underlines the need for dedicated imaging to accurately understand the mechanism of valve failure, and the feasibility of a valve-in-valve procedure. METHODS: A patient with structural bioprosthetic deterioration at echocardiography was investigated using computed tomography (CT) scanning and novel 3D Slicer software. The findings were compared with those revealed at intraoperative pathology. RESULTS: Post-processed CT images showed the bulk of calcifications to be located at the subvalvular level, suggesting the presence of calcified pannus. Pathology of the explanted valve confirmed that the valve stenosis was due primarily to pannus. Misdiagnosed calcified pannus represents a major threat during valve-in-valve procedures, due mainly to embolic risk. CONCLUSION: The 3D Slicer elaboration of CT images may be invaluable in providing a precise definition of the mechanism of valve failure, and also to establish the feasibility of either a valve-in-valve procedure or conventional surgery. |
|
Publication: J Neurol Surg A Cent Eur Neurosurg. 2013 Mar;74(2):71-6. PMID: 23404553 Authors: Inoue D, Cho B, Mori M, Kikkawa Y, Amano T, Nakamizo A, Yoshimoto K, Mizoguchi M, Tomikawa M, Hong J, Hashizume M, Sasaki T. Institution: Department of Neurosurgery, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan. Background/Purpose: To develop an augmented reality (AR) neuronavigation system with Web cameras and examine its clinical utility. The utility of the system was evaluated in three patients with brain tumors. One patient had a glioblastoma and two patients had convexity meningiomas. Our navigation system comprised the open-source software 3D Slicer (Brigham and Women's Hospital, Boston, Massachusetts, USA), the infrared optical tracking sensor Polaris (Northern Digital Inc., Waterloo, Canada), and Web cameras. We prepared two different types of Web cameras: a handheld type and a headband type. Optical markers were attached to each Web camera. We used this system for skin incision planning before the operation, during craniotomy, and after dural incision.Results We were able to overlay these images in all cases. In Case 1, accuracy could not be evaluated because the tumor was not on the surface, though it was generally suitable for the outline of the external ear and the skin. In Cases 2 and 3, the augmented reality error was ∼2 to 3 mm. AR technology was examined with Web cameras in neurosurgical operations. Our results suggest that this technology is clinically useful in neurosurgical procedures, particularly for brain tumors close to the brain surface. |
Can transrectal needle biopsy be optimised to detect nearly all prostate cancer with a volume of ≥0.5 mL? A three-dimensional analysis
|
Publication: BJU Int. 2013 Mar 12. PMID: 23490279 Authors: Kanao K, Eastham JA, Scardino PT, Reuter VE, Fine SW. Institution: Department of Pathology, Urology Service, Memorial Sloan-Kettering Cancer Center; Department of Surgery, Urology Service, Memorial Sloan-Kettering Cancer Center. Background/Purpose: Retrospectively analysed 109 whole-mounted and entirely submitted radical prostatectomy specimens with prostate cancer. All tumours in each prostate were outlined on whole-mount slides and digitally scanned to produce tumour maps. Tumour map images were exported to three-dimensional 3D Slicer software (http://www.slicer.org) to develop a 3D-prostate cancer model. In all, 20 transrectal biopsy schemes involving two to 40 cores and two to six anteriorly directed biopsy (ADBx) cores (including transition zone, TZ) were simulated, as well as models with various biopsy cutting lengths. Detection rates for tumours of different volumes were determined for the various biopsy simulation schemes. |
Quantitative MRI Analysis of Craniofacial Bone Marrow in Patients with Sickle Cell Disease
|
Publication: AJNR Am J Neuroradiol. 2013 Mar;34(3):622-7. PMID: 22878006 Authors: Elias EJ, Liao JH, Jara H, Watanabe M, Nadgir RN, Sakai Y, Erbay K, Saito N, Ozonoff A, Steinberg MH, Sakai O. Institutions:
Background/Purpose: Assessment of bone marrow is most commonly performed qualitatively in the spine or other large long bones. The craniofacial bones are less ideal for bone marrow analysis because of the relatively small bone marrow volume. Because patients with SCD often undergo repeated brain imaging to evaluate for cerebral vaso-occlusive disease, quantitative assessment of craniofacial bone marrow is a reasonable possibility in these patients. The purpose of this study was to investigate specific sickle cell disease changes in craniofacial bone marrow quantitatively by analyzing T1, T2, and secular-T2 relaxation times and volume with the use of quantitative MRI. Fourteen patients with SCD and 17 control subjects were imaged with the mixed TSE pulse sequence at 1.5T. The craniofacial bones were manually segmented by using 3D Slicer to generate bone marrow volumes and to provide T1, T2, and secular-T2 relaxation times. All subjects exhibited a bimodal T1 histogram. In the SCD group, there was a decrease in amplitude in the first T1 peak and an increase in amplitude in the second T1 peak. The first T1 peak showed a significant increase in relaxation time compared with control subjects (P < .0001), whereas there was no significant difference in the second T1 peak. T2 and secular-T2 relaxation times were significantly shorter in the SCD group (T2, P < .0001; secular-T2, P < .0001). Increasing numbers of blood transfusions resulted in a decrease in T2 and secular-T2 times. Patients with SCD exhibited a larger bone marrow volume compared with control subjects, even after standardization. Patients with SCD exhibited significant quantifiable changes in the craniofacial bone marrow because of failure of red-to-yellow marrow conversion and iron deposition that can be identified by qMRI relaxometry and volumetry. Both qMRI relaxometry and volumetry may be used as noninvasive tools for assessment of disease severity. |
Three-dimensional volumetric rendition of cannon ball pulmonary metastases by the use of 3D Slicer, an open source free software package
|
Publication: BMJ Case Rep. 2013 Feb 4;2013. PMID: 23386494 | PDF Authors: Revannasiddaiah S, Susheela SP, Madhusudhan N, Mallarajapatna GJ. Institution: Department of Radiation Oncology, HealthCare Global-Bangalore Institute of Oncology, Bengaluru, Karnataka, India. Background/Purpose: A gentleman treated 6 years ago for colonic carcinoma (stage-IIA) had been on regular follow-up for a span of 4 years, when he was apparently disease-free. After being lost to follow-up in the past 2 years, he was recently diagnosed with a pathological fracture of the seventh thoracic vertebra for which he underwent surgery (vertebral fixation and cord decompression) on an emergency basis. Postoperatively, he underwent a CT simulation of the thorax with the purpose of planning radiotherapy (RT) to the involved vertebrae. The CT-scan demonstrated multiple bilateral pulmonary nodules. After palliative RT to the involved vertebral column, in the interest of academics, we utilised the CT-scan data for volumetric reconstructions to attempt a better depiction of the well-known phenomenon of ‘cannon ball metastases’. This was achieved by the use of Slicer 4.2, an open source software package (distributed under a BSD-style open source license) intended for advanced visualisation and analysis of medical image datasets. This, to our knowledge, is possibly the first publication providing a three-dimensional depiction of ‘cannon ball’ pulmonary metastases. |
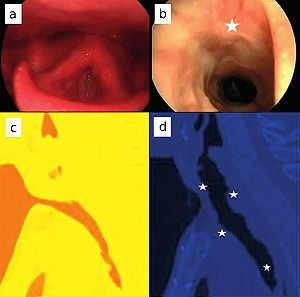
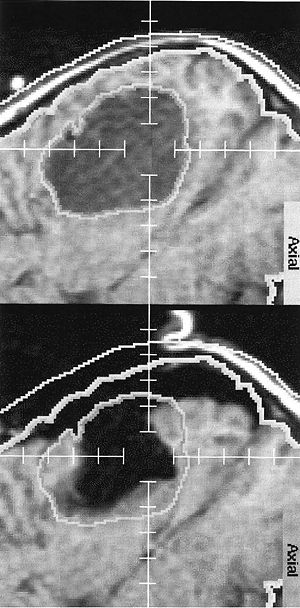
MRI Evidence for Altered Venous Drainage and Intracranial Compliance in Mild Traumatic Brain Injury
|
Publication: PLoS One. 2013 February;;8(2):e55447. PMID: 23405151 | PDF Authors: Pomschar A, Koerte I, Lee S, Laubender RP, Straube A, Heinen F, Ertl-Wagner B, Alperin N. Institution: Institute of Clinical Radiology, University of Munich - Grosshadern Campus, Ludwig-Maximilians-University Munich, Munich, Germany. Background/Purpose: To compare venous drainage patterns and associated intracranial hydrodynamics between subjects who experienced mild traumatic brain injury (mTBI) and age- and gender-matched controls. Thirty adult subjects (15 with mTBI and 15 age- and gender-matched controls) were investigated using a 3T MR scanner. Time since trauma was 0.5 to 29 years (mean 11.4 years). A 2D-time-of-flight MR-venography of the upper neck was performed to visualize the cervical venous vasculature. Cerebral venous drainage through primary and secondary channels, and intracranial compliance index and pressure were derived using cine-phase contrast imaging of the cerebral arterial inflow, venous outflow, and the craniospinal CSF flow. The intracranial compliance index is the defined as the ratio of maximal intracranial volume and pressure changes during the cardiac cycle. MR estimated ICP was then obtained through the inverse relationship between compliance and ICP. Funding:
|
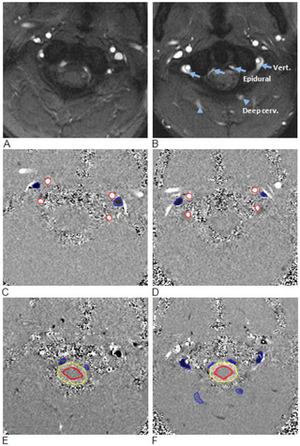 Examples of high and low velocity encoded phase contrast images from a control subject (left) and a subject with mTBI (right). A–B: Flow compensated magnitude images showing the bright signal from blood vessels. The augmented venous outflow through the epidural, vertebral veins (arrows) and the deep cervical veins (arrow heads) is well visualized. C–D: High-velocity encoding images used for measurements of arterial inflow and venous outflow through the jugular veins. E–F: Low-velocity encoding images used for measurements of the flow through the secondary channels (epidural, vertebral, and deep cervical veins) and the CSF flow. The lumen boundaries (red – arteries, blue- veins and yellow and red – CSF and cord) were identified using the PUBS automated segmentation method. |
Augmented reality visualisation using an image overlay system for MR-guided interventions: technical performance of spine injection procedures in human cadavers at 1.5 Tesla
|
Publication: Eur Radiol. 2013 Jan;23(1):235-45. PMID: 22797956 Authors: Fritz J, U-Thainual P, Ungi T, Flammang AJ, Fichtinger G, Iordachita II, Carrino JA Institution: Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, 601 North Caroline Street, Baltimore, MD, USA. Background/Purpose: To prospectively assess the technical performance of an augmented reality system for MR-guided spinal injection procedures. The augmented reality system was used with a clinical 1.5-T MRI system. A total of 187 lumbosacral spinal injection procedures (epidural injection, spinal nerve root injection, facet joint injection, medial branch block, discography) were performed in 12 human cadavers. Needle paths were planned with the Perk Station module of 3D Slicer software on high-resolution MR images. Needles were placed under augmented reality MRI navigation. MRI was used to confirm needle locations. T1-weighted fat-suppressed MRI was used to visualise the injectant. Outcome variables assessed were needle adjustment rate, inadvertent puncture of non-targeted structures, successful injection rate and procedure time. Needle access was achieved in 176/187 (94.1 %) targets, whereas 11/187 (5.9 %) were inaccessible. Six of 11 (54.5 %) L5-S1 disks were inaccessible, because of an axial obliquity of 30˚ (27˚-34˚); 5/11 (45.5 %) facet joints were inaccessible because of osteoarthritis or fusion. All accessible targets (176/187, 94.1 %) were successfully injected, requiring 47/176 (26.7 %) needle adjustments. There were no inadvertent punctures of vulnerable structures. Median procedure time was 10.2 min (5-19 min). Image overlay navigated MR-guided spinal injections were technically accurate. Disks with an obliquity ≥27˚ may be inaccessible. |
2012
Perk Tutor: an open-source training platform for ultrasound-guided needle insertions
|
Publication: IEEE Trans Biomed Eng. 2012 Dec;59(12):3475-81. PMID: 23008243 | PDF Authors: Ungi T, Sargent D, Moult E, Lasso A, Pinter C, McGraw RC, Fichtinger G. Institution: School of Computing, Queen's University, Kingston, ON, Canada. Background/Purpose: mage-guided needle placement, including ultrasound (US)-guided techniques, have become commonplace in modern medical diagnosis and therapy. To ensure that the next generations of physicians are competent using this technology, efficient and effective educational programs need to be developed. This paper presents the Perk Tutor: a configurable, open-source training platform for US-guided needle insertions. The Perk Tutor was successfully tested in three different configurations to demonstrate its adaptability to different procedures and learning objectives. 1) The Targeting Tutor, designed to develop US-guided needle targeting skills, 2) the Lumbar Tutor, designed for practicing US-guided lumbar spinal procedures, and (3) the Prostate Biopsy Tutor, configured for US-guided prostate biopsies. The Perk Tutor provides the trainee with quantitative feedback on progress toward the specific learning objectives of each configuration. Configurations were implemented through simple rearrangement of hardware and software components, attesting to the modularity and ease of configuration. The Perk Tutor is provided as a free resource to enable research and development of educational programs for US-guided intervention. |
Pituitary Adenoma Volumetry with 3D Slicer
|
Publication: PLoS One. 2012 Dec;7(12):e51788. PMID: 23240062 | PDF Authors: Egger J, Kapur T, Nimsky C, Kikinis R. Institution: Department of Radiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA. Background/Purpose: In this study, we present pituitary adenoma volumetry using the free and open source medical image computing platform for biomedical research: 3D Slicer. Volumetric changes in cerebral pathologies like pituitary adenomas are a critical factor in treatment decisions by physicians and in general the volume is acquired manually. Therefore, manual slice-by-slice segmentations in magnetic resonance imaging (MRI) data, which have been obtained at regular intervals, are performed. In contrast to this manual time consuming slice-by-slice segmentation process, 3D Slicer is an alternative which can be significantly faster and less user intensive. In this contribution, we compare pure manual segmentations of ten pituitary adenomas with semi-automatic segmentations with 3D Slicer. Thus, physicians drew the boundaries completely manually on a slice-by-slice basis and performed a Slicer-enhanced segmentation using the competitive region-growing based module of 3D Slicer named GrowCut. Results showed that the time and user effort required for GrowCut-based segmentations were on average about thirty percent less than the pure manual segmentations. Furthermore, we calculated the Dice Similarity Coefficient (DSC) between the manual and the Slicer-based segmentations to proof that the two are comparable yielding an average DSC of 81.9763.39%. |
Differences in simple morphological variables in ruptured and unruptured middle cerebral artery aneurysms
|
Publication: J Neurosurg. 2012 Nov;117(5):913-9. PMID: 22957531 Authors: Lin N, Ho A, Gross BA, Pieper S, Frerichs KU, Day AL, Du R. Institution: Department of Neurosurgery, and Surgical Planning Laboratory, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA. Background/Purpose: Object Management of unruptured intracranial aneurysms remains controversial in neurosurgery. The contribution of morphological parameters has not been included in the treatment paradigm in a systematic manner or for any particular aneurysm location. The authors present a large sample of middle cerebral artery (MCA) aneurysms that were assessed using morphological variables to determine the parameters associated with aneurysm rupture. Methods Preoperative CT angiography (CTA) studies were evaluated using 3D Slicer software to generate 3D models of the aneurysms and their surrounding vascular architecture. Morphological parameters examined in each model included 5 variables already defined in the literature (aneurysm size, aspect ratio, aneurysm angle, vessel angle, and size ratio) and 3 novel variables (flow angle, distance to the genu, and parent-daughter angle). Univariate and multivariate statistical analyses were performed to determine statistical significance. Results Between 2005 and 2008, 132 MCA aneurysms were treated at a single institution, and CTA studies of 79 aneurysms (40 ruptured and 39 unruptured) were analyzed. Fifty-three aneurysms were excluded because of reoperation (4), associated AVM (2), or lack of preoperative CTA studies (47). Ruptured aneurysms were associated with larger size, greater aspect ratio, larger aneurysm and flow angles, and smaller parent-daughter angle. Multivariate logistic regression revealed that aspect ratio, flow angle, and parent-daughter angle were the strongest factors associated with ruptured aneurysms. Aspect ratio, flow angle, and parent-daughter angle are more strongly associated with ruptured MCA aneurysms than size. The association of parameters independent of aneurysm morphology with ruptured aneurysms suggests that these parameters may be associated with an increased risk of aneurysm rupture. These factors are readily applied in clinical practice and should be considered in addition to aneurysm size when assessing the risk of aneurysm rupture specific to the MCA location. |
3D Slicer as an image computing platform for the Quantitative Imaging Network
|
Publication: Magn Reson Imaging. 2012 Nov;30(9):1323-41. PMID: 22770690 | PDF Authors: Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R. Institution: Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA. Background/Purpose: Quantitative analysis has tremendous but mostly unrealized potential in healthcare to support objective and accurate interpretation of the clinical imaging. In 2008, the National Cancer Institute began building the Quantitative Imaging Network (QIN) initiative with the goal of advancing quantitative imaging in the context of personalized therapy and evaluation of treatment response. Computerized analysis is an important component contributing to reproducibility and efficiency of the quantitative imaging techniques. The success of quantitative imaging is contingent on robust analysis methods and software tools to bring these methods from bench to bedside. 3D Slicer is a free open-source software application for medical image computing. As a clinical research tool, 3D Slicer is similar to a radiology workstation that supports versatile visualizations but also provides advanced functionality such as automated segmentation and registration for a variety of application domains. Unlike a typical radiology workstation, 3D Slicer is free and is not tied to specific hardware. As a programming platform, 3D Slicer facilitates translation and evaluation of the new quantitative methods by allowing the biomedical researcher to focus on the implementation of the algorithm and providing abstractions for the common tasks of data communication, visualization and user interface development. Compared to other tools that provide aspects of this functionality, 3D Slicer is fully open source and can be readily extended and redistributed. In addition, 3D Slicer is designed to facilitate the development of new functionality in the form of 3D Slicer extensions. In this paper, we present an overview of 3D Slicer as a platform for prototyping, development and evaluation of image analysis tools for clinical research applications. To illustrate the utility of the platform in the scope of QIN, we discuss several use cases of 3D Slicer by the existing QIN teams, and we elaborate on the future directions that can further facilitate development and validation of imaging biomarkers using 3D Slicer. |
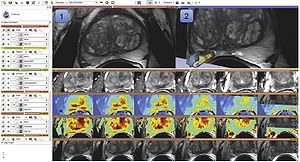 Visualization capabilities and 3D Slicer user interface are utilized for exploration of the multiparametric MRI of the prostate. The T2w MR scans (top left image and the bottom row of consecutive slices) provide anatomical detail and context for interpretation of the registered ADC map (first row of the image matrix) calculated from the DWI and various pharmacokinetic maps computed from DCE MRI, as shown in the 2D axial slice viewer (marked by “1”) and the Compare View layout below. Spatial context of the annotated tumor ROI is provided by composed view of the tumor ROI surface and the T2w image cross-section in the 3D viewer (marked by “2”). |
SlicerRT: radiation therapy research toolkit for 3D Slicer
|
Publication: Med Phys. 2012 Oct;39(10):6332-8. PMID: 23039669 | PDF Authors: Pinter C, Lasso A, Wang A, Jaffray D, Fichtinger G. Institution: School of Computing, Queen's University, Kingston, Ontario, Canada. Background/Purpose: Interest in adaptive radiation therapy research is constantly growing, but software tools available for researchers are mostly either expensive, closed proprietary applications, or free open-source packages with limited scope, extensibility, reliability, or user support. To address these limitations, we propose SlicerRT, a customizable, free, and open-source radiation therapy research toolkit. SlicerRT aspires to be an open-source toolkit for RT research, providing fast computations, convenient workflows for researchers, and a general image-guided therapy infrastructure to assist clinical translation of experimental therapeutic approaches. It is a medium into which RT researchers can integrate their methods and algorithms, and conduct comparative testing. SlicerRT was implemented as an extension for the widely used 3D Slicer medical image visualization and analysis application platform. SlicerRT provides functionality specifically designed for radiation therapy research, in addition to the powerful tools that 3D Slicer offers for visualization, registration, segmentation, and data management. The feature set of SlicerRT was defined through consensus discussions with a large pool of RT researchers, including both radiation oncologists and medical physicists. The development processes used were similar to those of 3D Slicer to ensure software quality. Standardized mechanisms of 3D Slicer were applied for documentation, distribution, and user support. The testing and validation environment was configured to automatically launch a regression test upon each software change and to perform comparison with ground truth results provided by other RT applications. Modules have been created for importing and loading DICOM-RT data, computing and displaying dose volume histograms, creating accumulated dose volumes, comparing dose volumes, and visualizing isodose lines and surfaces. The effectiveness of using 3D Slicer with the proposed SlicerRT extension for radiation therapy research was demonstrated on multiple use cases. A new open-source software toolkit has been developed for radiation therapy research. SlicerRT can import treatment plans from various sources into 3D Slicer for visualization, analysis, comparison, and processing. The provided algorithms are extensively tested and they are accessible through a convenient graphical user interface as well as a flexible application programming interface. |
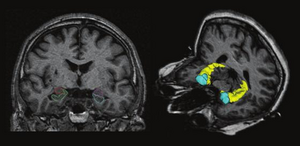
Developmental Trajectories of Amygdala and Hippocampus from Infancy to Early Adulthood in Healthy Individuals
|
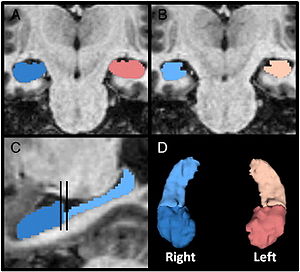
Publication: PLoS One. 2012 October;7(10):e46970. PMID: 23056545 | PDF Authors: Uematsu A, Matsui M, Tanaka C, Takahashi T, Noguchi K, Suzuki M, Nishijo H. Institution: Department of Psychology, Graduate School of Medicine and Pharmaceutical Sciences, University of Toyama, Toyama, Japan. Background/Purpose: Knowledge of amygdalar and hippocampal development as they pertain to sex differences and laterality would help to understand not only brain development but also the relationship between brain volume and brain functions. However, few studies investigated development of these two regions, especially during infancy. The purpose of this study was to examine typical volumetric trajectories of amygdala and hippocampus from infancy to early adulthood by predicting sexual dimorphism and laterality. We performed a cross-sectional morphometric MRI study of amygdalar and hippocampal growth from 1 month to 25 years old, using 109 healthy individuals. The findings indicated significant non-linear age-related volume changes, especially during the first few years of life, in both the amygdala and hippocampus regardless of sex. The peak ages of amygdalar and hippocampal volumes came at the timing of preadolescence (9-11 years old). The female amygdala reached its peak age about one year and a half earlier than the male amygdala did. In addition, its rate of growth change decreased earlier in the females. Furthermore, both females and males displayed rightward laterality in the hippocampus, but only the males in the amygdala. The robust growth of the amygdala and hippocampus during infancy highlight the importance of this period for neural and functional development. The sex differences and laterality during development of these two regions suggest that sex-related factors such as sex hormones and functional laterality might affect brain development. Funding:
|
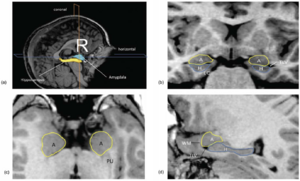 Orthogonal views for segmenting the amygdala and hippocampus on MRI sections. A three-dimensional reconstruction of images (a) in which lines indicate the position of the horizontal plane (b), sagittal plane (c), and coronal plane (d) is shown. A, Amygdala; EC, entorhinal cortex; H, hippocampus; PU, putamen; TLV, temporal horn of the lateral ventricle; WM, subamygdaloid white matter. For the amygdala and hippocampus volumes, the images were imported into the 3D Slicer software (http://www.slicer.org/) and manually defined. |
Extra-Hippocampal Subcortical Limbic Involvement Predicts Episodic Recall Performance in Multiple Sclerosis
|
Publication: PLoS One. 2012 October;7(10):e44942. PMID: 23056187 | PDF Authors: Dineen RA, Bradshaw CM, Constantinescu CS, Auer DP. Institution: Division of Radiological and Imaging Sciences, University of Nottingham, Nottingham, UK. Background/Purpose: Episodic memory impairment is a common but poorly-understood phenomenon in multiple sclerosis (MS). We aim to establish the relative contributions of reduced integrity of components of the extended hippocampal-diencephalic system to memory performance in MS patients using quantitative neuroimaging. 34 patients with relapsing-remitting MS and 24 healthy age-matched controls underwent 3 T MRI including diffusion tensor imaging and 3-D T1-weighted volume acquisition. Manual fornix regions-of-interest were used to derive fornix fractional anisotropy (FA). Normalized hippocampal, mammillary body and thalamic volumes were derived by manual segmentation. MS subjects underwent visual recall, verbal recall, verbal recognition and verbal fluency assessment. Significant differences between MS patients and controls were found for fornix FA (0.38 vs. 0.46, means adjusted for age and fornix volume, P<.0005) and mammillary body volumes (age-adjusted means 0.114 ml vs. 0.126 ml, P<.023). Multivariate regression analysis identified fornix FA and mammillary bodies as predictor of visual recall (R(2) = .31, P = .003, P = .006), and thalamic volume as predictive of verbal recall (R(2) = .37, P<.0005). No limbic measures predicted verbal recognition or verbal fluency. These findings indicate that structural and ultrastructural alterations in subcortical limbic components beyond the hippocampus predict performance of episodic recall in MS patients with mild memory dysfunction. Funding:
|
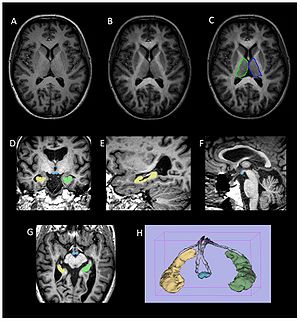 Volumetric segmentation of brain images. (a–c) Thalamic segmentation. Axial MPRAGE image from a control subject (a) before and (b) after the addition of FA weighting from the co-registered FA image. Note the increase in clarity of the lateral border of the thalamus that facilitates (c) manual segmentation of the thalamus. (d–g) axial, coronal and sagittal images from a participant with MS demonstrating ROIs drawn for the mammillary bodies (blue) and right (yellow) and left (green) hippocampi. (h) Surface rendered representation of the segmented hippocampi and mammillary bodies from a participant with MS. The manually defined fornix ROI used to derive fornix FA is also shown (grey). Volumes of the hippocampi, mammillary bodies and thalami were obtained manually from the MPRAGE images using 3D Slicer software |
Posterior vaginal prolapse shape and position changes at maximal Valsalva seen in 3-D MRI-based models.
|
Publication: Int Urogynecol J. 2012 Sep;23(9):1301-6. PMID: 22527556 Authors: Luo J, Larson KA, Fenner DE, Ashton-Miller JA, Delancey JO. Institution: Pelvic Floor Research Group, University of Michigan, Ann Arbor, MI, USA. Background/Purpose: Two-dimensional magnetic resonance imaging (MRI) of posterior vaginal prolapse has been studied. However, the three-dimensional (3-D) mechanisms causing such prolapse remain poorly understood. This discovery project was undertaken to identify the different 3-D characteristics of models of rectocele-type posterior vaginal prolapse (PVP(R)) in women. Ten women with (cases) and ten without (controls) PVP(R) were selected from an ongoing case-control study. Supine, multiplanar MR imaging was performed at rest and maximal Valsalva. Three-dimensional reconstructions of the posterior vaginal wall and pelvic bones were created using 3D Slicer v. 3.4.1. In each slice the posterior vaginal wall and perineal skin were outlined to the anterior margin of the external anal sphincter to include the area of the perineal body. Women with predominant enteroceles or anterior vaginal prolapse were excluded. The case and control groups had similar demographics. In women with PVP(R) two characteristics were consistently visible (10/10): (1) the posterior vaginal wall displayed a folding phenomenon similar to a person beginning to kneel ("kneeling" shape) and (2) a downward displacement in the upper two thirds of the vagina. Also seen in some, but not all of the scans were: (3) forward protrusion of the distal vagina (6/10), (4) perineal descent (5/10), and (5) distal widening in the lower third of the vagina (3/10). Increased folding (kneeling) of the vagina and an overall downward displacement are consistently present in rectocele. Forward protrusion, perineal descent, and distal widening are sometimes seen as well. |
Talairach methodology in the multimodal imaging and robotics era
|
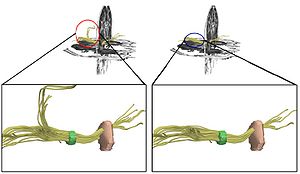
Publication: Book Chapter in: Jean-Marie Scarabin Ed. Stereotaxy and Epilepsy Surgery. John Libbey Eurotext LTD. 2012 Sept; p.245-272. ISBN-10: 2742007679. PDF Authors: Cardinale F, Miserocchi A, Moscato A, Cossu M, Castana L, Schiariti M P, Gozzo F, Pero G, Quilici L, Citterio A, Minella M, Torresin A, Lo Russo G. Institution: Centro Claudio Munari per la Chirurgica dell' Epilessia e del Parkinson, Ospedale Niguarda Ca' Grande, Milan, Italy. Background/Purpose: The original Talairach methodology was based on three main features. Neuroimaging techniques to collect patient-specific brain anatomical data, a probabilistic morpho-functional atlas and surgical tools developed to implant intracerebral electrodes in accurate and safe conditions. Brain anatomy was obtained indirectly by means of cerebral angiography and ventriculography. Both techniques were performed in stereotactic (coordinate system), telemetric (reduced magnification and distortion) and stereoscopic (pseudo-3D effect) conditions. The following step was to normalize patient’s specific data to the atlas space using the bicommissural coordinate system. The normalization to the atlas was fundamental in order to integrate the images with structural (gyri and sulci), cytoarchitectural (Brodmann areas) and functional (electrical stimulations) data. Then, in order to position the guiding screws along the pre-planned trajectory, a double grid was attached onto the frame, allowing drilling in the orthogonal direction. Nowadays modern technological tools facilitate the workflow, but the main concepts of this methodology are still valid. We use algorithms completely automated with software applications to expand the multimodal integration of data so that MRI, rotational angiography, fMRI and DTI-FT data can all be coregistered together. It is now possible to obtain real 3D viewing by multi-planar reconstructions (MPR), volume and surface rendering. The tool holder for the surgical implantation is now fixed onto the passive robotic arm of the stereotactic imageguided system Renishaw neuromate®, so that the number of possible trajectories is no more discrete (as using the double grid), but virtually infinite with any desired obliquity. The aim of this chapter is to describe our present workflow. We will underline the methodological continuity with basic Talairach and Bancaud principles, highlighting how modern tools can help in saving time and improve safety, accuracy and the amount of information. Moreover, we report our quantitative results regarding geometrical accuracy at the cortical entry point with this new methodology. |
 3D Slicer scene with MPR and 3D views (crossedviewing)During long-term Video-SEEGmonitoring, for several daysfollowing implantation, it isimportant to have intuitive andaccurate mapping tools for localizing the sources of the signal. In order to reach this aim, we integrate all the information in 3D Slicer, a package able to load all data. It is in fact possible to import all datasets and surface models computed by FreeSurfer (white and gray matter, with parcellations and labels). Moreover, it is possible to create other models such as the vascular tree and the electrodes leads. This is done very simply with the Gray Scale Model Maker tool. 3D Slicer is installed on all team members’ computers, so that epileptologists are able to look at these scenes when interpreting the SEEG traces. (A and B) constitute a pair of stereoscopic images with the reconstruction of pial surface, vessels and labeled electrodes. (C, D and E) are the MPR views, with the visualization fading 50% for MRI and 50% forpost-implant O-arm dataset. The crosshair indicates thesecond lead of an electrode exploring first temporal gyrus,temporal transverse gyri and insular long gyri. |
SARRP Dose Planning System
|
Company: Xstrahl Ltd., Camberley, United Kingdom. Background: The SARRP dose planning system (DPS) is a dedicated in vivo treatment planning system designed to calculate dose distributions in pre-clinical radiation studies. The SARRP DPS is specifically validated for use with in vivo—uniquely enabling the user to treat very precise areas of the animal by defining the area and visualizing the dose and as a result, enabling a more accurate experiment and final result. More... |
Three-dimensional image quantification as a new morphometry method for microvascular tissue engineering
|
Publication: Tissue Eng Part C Methods. 2012 Jul;18(7):507-16. PMID: 22224751 Authors: Rytlewski JA, Geuss LR, Anyaeji CI, Lewis EW, Suggs L. Institution: The University of Texas at Austin, Biomedical Engineering, Austin, TX, USA. Background/Purpose: Morphological analysis is an essential step in verifying the success of a tissue engineering strategy where researchers must determine the presence of a desired cellular phenotype. While morphometry has transitioned from observational grading to computational quantification, established quantitative methods eliminate information by relying on two-dimensional (2D) analysis to describe three-dimensional (3D) niches. In this study, we demonstrate the validity and utility of 3D morphological quantification using two common angiogenesis assays in our fibrin-based in vitro model: (1) the microcarrier bead assay with human mesenchymal stem cells and (2) the rat aortic ring outgrowth assay. The quantification method is based on collecting and segmenting fluorescent confocal z-stacks into 3D models with 3D Slicer, an open-source MRI/CT analysis program. Data from 3D models are then processed into biologically relevant metrics in MATLAB for statistical analysis. Metrics include descriptive parameters such as vascular network length, volume, number of network segments, and degree of network branching. Our results indicate that 2D measures are significantly different than their 3D counterparts unless the vascular network exhibits anisotropic growth along the plane of imaging. Additionally, the statistical outcomes of 3D morphological quantification agreed with our initial qualitative observations among different test groups. This novel quantification approach generates more spatially accurate and objective measures, representing an important step towards improving the reliability of morphological comparisons. |
Prostate: registration of digital histopathologic images to in vivo MR images acquired by using endorectal receive coil
|
Publication: Radiology. 2012 Jun;263(3):856-64 PMID: 22474671 | PDF Authors: Ward AD, Crukley C, McKenzie CA, Montreuil J, Gibson E, Romagnoli C, Gomez JA, Moussa M, Chin J, Bauman G, Fenster A. Institution: Imaging Research Laboratories, Robarts Research Institute, Schulich School of Medicine and Dentistry, the University of Western Ontario, London, ON, Canada. Background/Purpose: To develop and evaluate a technique for the registration of in vivo prostate magnetic resonance (MR) images to digital histopathologic images by using image-guided specimen slicing based on strand-shaped fiducial markers relating specimen imaging to histopathologic examination.The study was approved by the institutional review board (the University of Western Ontario Health Sciences Research Ethics Board, London, Ontario, Canada), and written informed consent was obtained from all patients. This work proposed and evaluated a technique utilizing developed fiducial markers and real-time three-dimensional visualization in support of image guidance for ex vivo prostate specimen slicing parallel to the MR imaging planes prior to digitization, simplifying the registration process. The registration was performed interactively by using 3D Slicer (Surgical Planning Laboratory, Harvard Medical School, Boston, MA)Means, standard deviations, root-mean-square errors, and 95% confidence intervals are reported for all evaluated measurements. The slicing error was within the 2.2 mm thickness of the diagnostic-quality MR imaging sections, with a tissue block thickness standard deviation of 0.2 mm. Rigid registration provided negligible postregistration overlap of the smallest clinically important tumors (0.2 cm3) at histologic examination and MR imaging, whereas the tested nonrigid registration method yielded a mean target registration error of 1.1 mm and provided useful coregistration of such tumors. This method for the registration of prostate digital histo-pathologic images to in vivo MR images acquired by using an endorectal receive coil was sufficiently accurate for coregistering the smallest clinically important lesions with 95% confidence. |
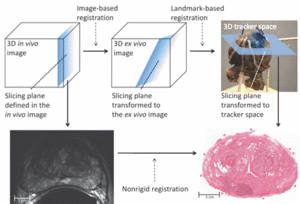 Image-guided specimen slicing process. The first step is to select a desired slicing plane orientation and location from the 3D in vivo image. By using image-based registration, this plane is transformed to the space of the ex vivo specimen image. Corresponding fiducial markers are localized on the ex vivo image (by using software) and on the physical specimen (by using a tracked stylus), and a landmark transform aligning these fiducial markers transforms the slicing plane into the space of the tracker. The tracked stylus is used to direct the insertion of three slicing plane–defining pins into the specimen, which align the specimen in a slotted forceps for slicing. This permits the establishment of a correspondence between stained histologic slices and in vivo imaging planes, which are registered by using a nonrigid registration. |
A comparison of the effect of age on levator ani and obturator internus muscle cross-sectional areas and volumes in nulliparous women
|
Publication: Neurourol Urodyn. 2012 Apr;31(4):481-6. PMID: 22378544 Authors: Morris VC, Murray MP, Delancey JO, Ashton-Miller JA. Institution: University College London Hospital, London, England. Background/Purpose: Functional tests have demonstrated minimal loss of vaginal closure force with age. So we tested the null hypotheses that age neither affects the maximum cross-sectional area (CSA) nor the volume of the levator muscle. Corresponding hypotheses were also tested in the adjacent obturator internus muscle, which served as a control for the effect of age on appendicular muscle in these women. Magnetic resonance images of 15 healthy younger (aged 21-25 years) and 12 healthy older nulliparous women (aged >63 years) were selected to avoid the confounding effect of childbirth. Models were created from tracing outlines of the levator ani muscle in the coronal plane, and obturator internus in the axial plane using 3D Slicer v. 3.4. Muscle volumes were calculated using 3D Slicer, while CSA was measured using Imageware™ at nine locations. The hypotheses were tested using repeated measures analysis of variance with P < 0.05 being considered significant. The effect of age did not reach statistical significance for the decrease in levator ani muscle maximum CSA or the decrease in volume (4.3%, P = 0.62 and 10.9%, 0.12, respectively). However, age did significantly adversely decrease obturator internus muscle maximum CSA and volume (24.5% and 28.2%, P < 0.001, respectively). Significant local age-related changes were observed dorsally in both muscles. Unlike the adjacent appendicular muscle, obturator internus, the levator ani muscle in healthy nullipara does not show evidence of significant age-related atrophy. Funding:
|
Levator ani subtended volume: a novel parameter to evaluate levator ani muscle laxity in pelvic organ prolapse
|
Publication: Am J Obstet Gynecol. 2012 Mar;206(3):244.e1-9. PMID: 22075059 | PDF Authors: Rodrigues AA Jr, Bassaly R, McCullough M, Terwilliger HL, Hart S, Downes K, Hoyte L. Institution: Division of Female Pelvic Medicine and Reconstructive Surgery, Department of Obstetrics and Gynecology, University of South Florida College of Medicine, Tampa, FL. Background/Purpose: We describe a new parameter based on magnetic resonance 3-dimensional (3D) reconstructions proposed to evaluate levator ani muscle (LAM) laxity in women with pelvic organ prolapse (POP). This is an institutional review board-approved, retrospective chart review of 35 women with POP, stages I-IV. The 3D Slicer software package was used to perform 2-dimensional and 3D measurements and the levator ani subtended volume (LASV) was described. Basically, the LASV represents the volume contained by LAM between 2 planes, which coincides with pubococcygeal line and H line. Correlations among measurements, ordinal POP stages, POP Quantification (POPQ) individual measurements, and validated questionnaires were performed. The LASV differentiated major (III and IV) from minor (I and II) POPQ stages, which positively correlated to POP stages and POPQ individual measurements. The LASV is a promising parameter to evaluate the LAM laxity. |
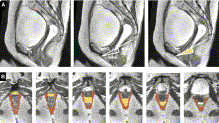 Two sequences showing segmentation processA, Midsagittal views of pelvic floor; representations of pubococcygeal line (PCL), H line, and M line (white); and anterior (yellow) and posterior (green) limits of levator hiatus volume. B, Levator ani muscle (LAM) and levator hiatus segmentation following caudal-cranial orientation. Levator hiatus was segmented based on previous limits obtained from midsagittal slice and inner boundaries of LAM, obturator internus muscle, and pubic bone.Rodrigues. A new parameter proposed to evaluate the levator ani laxity. |
Tractography Delineates Microstructural Changes in the Trigeminal Nerve after Focal Radiosurgery for Trigeminal Neuralgia
|
Publication: PLoS One. 2012 March;7(3):e32745. PMID:22412918 | PDF Authors: Hodaie M, Chen DQ, Quan J, Laperriere N. Institution: Division of Neurosurgery, Toronto Western Hospital and University of Toronto, Toronto, Ontario, Canada. Background/Purpose: Focal radiosurgery is a common treatment modality for trigeminal neuralgia (TN), a neuropathic facial pain condition. Assessment of treatment effectiveness is primarily clinical, given the paucity of investigational tools to assess trigeminal nerve changes. Since diffusion tensor imaging (DTI) provides information on white matter microstructure, we explored the feasibility of trigeminal nerve tractography and assessment of DTI parameters to study microstructural changes after treatment. We hypothesized that trigeminal tractography provides more information than 2D-MR imaging, allowing detection of unique, focal changes in the target area after radiosurgery. Changes in specific diffusivities may provide insight into the mechanism of action of radiosurgery on the trigeminal nerve. Funding:
|
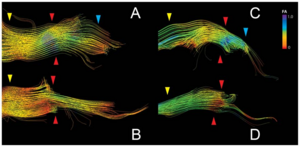 Tractography outlines detailed FA changes in the trigeminal nerve after GKRS treatment. Panels A–D depict the trigeminal nerve tracts pre and post-treatment for subjects S1(A,B) and S2 (C,D). The area between the yellow and blue arrows delineates the cisternal segment, with the yellow arrow being proximal to the brainstem and the blue arrow distal. The red arrow denotes the target area, which corresponds to the region where the greatest change in FA was observed. In S1, FA change affects primarily the outlying fibers of the nerve, while for S2, FA changes are seen in the inferior portion of the cisternal segment of the trigeminal nerve. Tractography were performed using the 3D Slicer software. |
Square-cut: A Segmentation Algorithm on the Basis of a Rectangle Shape
|
Publication: PLoS One. 2012 Feb;7(2):e31064. PMID: 22363547 | PDF Authors: Egger J, Kapur T, Dukatz T, Kolodziej M, Zukić D, Freisleben B, Nimsky C. Institution: Surgical Planning Laboratory, Department of Radiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA. Background/Purpose: We present a rectangle-based segmentation algorithm that sets up a graph and performs a graph cut to separate an object from the background. However, graph-based algorithms distribute the graph's nodes uniformly and equidistantly on the image. Then, a smoothness term is added to force the cut to prefer a particular shape. This strategy does not allow the cut to prefer a certain structure, especially when areas of the object are indistinguishable from the background. We solve this problem by referring to a rectangle shape of the object when sampling the graph nodes, i.e., the nodes are distributed non-uniformly and non-equidistantly on the image. This strategy can be useful, when areas of the object are indistinguishable from the background. For evaluation, we focus on vertebrae images from Magnetic Resonance Imaging (MRI) datasets to support the time consuming manual slice-by-slice segmentation performed by physicians. The ground truth of the vertebrae boundaries were manually extracted by two clinical experts (neurological surgeons) with several years of experience in spine surgery and afterwards compared with the automatic segmentation results of the proposed scheme yielding an average Dice Similarity Coefficient (DSC) of 90.97±2.2%. Funding:
|
Anatomical and functional correlates of human hippocampal volume asymmetry
|
Publication: Psychiatry Res. 2012 Jan; 201(1): 48–53. PMID: 22285719 | PDF Authors: Woolard AA, Heckers S. Institution: Department of Osychiatry, Vaderbilt University, Nashville, TN, USA. Background/Purpose: Hemispheric asymmetry of the human hippocampus is well established, but poorly understood. We studied 110 healthy subjects with 3-Tesla MRI to explore the anatomical and functional correlates of the R>L volume asymmetry. We found that the asymmetry is limited to the anterior hippocampus (hemisphere×region interaction: F(1,109)=42.6, p<.001). Anterior hippocampal volume was correlated strongly with the volumes of all four cortical lobes. In contrast, posterior hippocampal volume was correlated strongly only with occipital lobe volume, moderately with the parietal and temporal lobe volumes and not with the frontal lobe volume. The degree of R>L anterior hippocampal volume asymmetry predicted performance on a measure of basic cognitive abilities. This provides evidence for regional specificity and functional implications of the well-known hemispheric asymmetry of hippocampal volume. We suggest that the developmental profile, genetic mechanisms and functional implications of R>L anterior hippocampal volume asymmetry in the human brain deserve further study. |
Hippocampal subregions are differentially affected in the progression to Alzheimer's disease
|
Publication: Anat Rec (Hoboken). 2012 Jan;295(1):132-40. PMID: 22095921 PDF Authors: Greene SJ, Killiany RJ; Alzheimer's Disease Neuroimaging Initiative Institution: Department of Anatomy and Neurobiology, University of Vermont College of Medicine, Burlington, VT, USA. Background/Purpose: Atrophy within the hippocampus (HP) as measured by magnetic resonance imaging (MRI) is a promising biomarker for the progression to Alzheimer's disease (AD). Subregions of the HP along the longitudinal axis have been found to demonstrate unique function, as well as undergo differential changes in the progression to AD. Little is known of relationships between such HP subregions and other potential biomarkers, such as neuropsychological (NP), genetic, and cerebral spinal fluid (CSF) beta amyloid and tau measures. The purpose of this study was to subdivide the hippocampus to determine how the head, body, and tail were affected in normal control, mild cognitively impaired, and AD subjects, and investigate relationships with HP subregions and other potential biomarkers. MRI scans of 120 participants of the Alzheimer's Disease Neuroimaging Initiative were processed using FreeSurfer, and the HP was subdivided using 3D Slicer. Each subregion was compared among groups, and correlations were used to determine relationships with NP, genetic, and CSF measures. Results suggest that HP subregions are undergoing differential atrophy in AD, and demonstrate unique relationships with NP and CSF data. Discriminant function analyses revealed that these regions, when combined with NP and CSF measures, were able to classify by diagnostic group, and classify MCI subjects who would and would not progress to AD within 12 months. Funding:
|
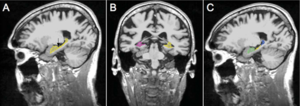 Methods for identifying boundaries for the head, body, and tail of the hippocampus. A: Arrow is pointing to the head/body boundary. B: View of body/tail boundary in left hemisphere where the fimbria of the fornix comes into view; body/tail boundary. C: Subregions after relabeling has been completed (green: head; yellow: body; blue: tail). |
Long-term memory search across the visual brain
|
Publication: Neural Plast. 2012;2012:392695. PMID: 22900206 | PDF Authors: Fedurco M. Institution: Michelin Recherche Technique, S.A., Fribourg, Switzerland. Background/Purpose: Signal transmission from the human retina to visual cortex and connectivity of visual brain areas are relatively well understood. How specific visual perceptions transform into corresponding long-term memories remains unknown. Here, I will review recent Blood Oxygenation Level-Dependent functional Magnetic Resonance Imaging (BOLD fMRI) in humans together with molecular biology studies (animal models) aiming to understand how the retinal image gets transformed into so-called visual (retinotropic) maps. The broken object paradigm has been chosen in order to illustrate the complexity of multisensory perception of simple objects subject to visual--rather than semantic--type of memory encoding. The author explores how amygdala projections to the visual cortex affect the memory formation and proposes the choice of experimental techniques needed to explain our massive visual memory capacity. Maintenance of the visual long-term memories is suggested to require recycling of GluR2-containing α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPAR) and β(2)-adrenoreceptors at the postsynaptic membrane, which critically depends on the catalytic activity of the N-ethylmaleimide-sensitive factor (NSF) and protein kinase PKMζ. |
 Parcellation of the human visual cortex and brain areas involved in visual object perception: (a) eye bulb—grey, optical tract/optical chiasm—white, lateral geniculate nucleus (LGN)—green, occipital visual cortex (OC)—white, lateral occipital gyrus (LOC)—navy blue, inferior temporal gyrus (ITC)—creamy yellow, fusiform gyrus (FG)—magenta, hippocampus (H)—red, amygdala (A)—cyan, thalamus (T)—grey (created using the 3D Slicer [38]). (b) Ventral occipital visual field map models: Cal-S (V1)—calcarine sulcus, V2—green, V3—blue, V4 (brick red), FG—fusiform gyrus, CoS—collateral sulcus, POS—parietal occipital sulcus. The dotted yellow line and white circles denote the region of cortex in which the V4 and V8 models diverge (inset shows inflated model of the left human hemisphere). (c) Anterior part of the fusiform gyrus is located in the vicinity of the hippocampal formation. (b) reproduced, with permission, from [25] Macmillan Publishers Ltd. (c) (right) reproduced, with permission, from [39] the American Physiological Society. |
Temporal course of cerebrospinal fluid dynamics and amyloid accumulation in the aging rat brain from three to thirty months
|
Publication: Fluids Barriers CNS. 2012 Jan 23;9(1):3. PMID: 22269091 | PDF Authors: Chiu C, Miller MC, Caralopoulos IN, Worden MS, Brinker T, Gordon ZN, Johanson CE, Silverberg GD. Institution: Department of Neurosurgery, Warren Alpert Medical School, Brown University and Aldrich Neurosurgery Research Laboratories, Rhode Island Hospital, Providence, RI, USA. Background/Purpose: Amyloid accumulation in the brain parenchyma is a hallmark of Alzheimer's disease (AD) and is seen in normal aging. Alterations in cerebrospinal fluid (CSF) dynamics are also associated with normal aging and AD. This study analyzed CSF volume, production and turnover rate in relation to amyloid-beta peptide (Aβ) accumulation in the aging rat brain. Aging Fischer 344/Brown-Norway hybrid rats at 3, 12, 20, and 30 months were studied. CSF production was measured by ventriculo-cisternal perfusion with blue dextran in artificial CSF; CSF volume by MRI; and CSF turnover rate by dividing the CSF production rate by the volume of the CSF space. Aβ40 and Aβ42 concentrations in the cortex and hippocampus were measured by ELISA. There was a significant linear increase in total cranial CSF volume with age: 3-20 months (p < 0.01); 3-30 months (p < 0.001). CSF production rate increased from 3-12 months (p < 0.01) and decreased from 12-30 months (p < 0.05). CSF turnover showed an initial increase from 3 months (9.40 day-1) to 12 months (11.30 day-1) and then a decrease to 20 months (10.23 day-1) and 30 months (6.62 day-1). Aβ40 and Aβ42 concentrations in brain increased from 3-30 months (p < 0.001). Both Aβ42 and Aβ40 concentrations approached a steady state level by 30 months. In young rats there is no correlation between CSF turnover and Aβ brain concentrations. After 12 months, CSF turnover decreases as brain Aβ continues to accumulate. This decrease in CSF turnover rate may be one of several clearance pathway alterations that influence age-related accumulation of brain amyloid. Fundings:
|
 Total CSF volume. A) CSF in the ventricles was measured by placing fiducial points (pink dots) on black pixels within the region of interest. FastMarching Segmentation in 3D Slicer generates a rough sketch of the region of interest (green areas), as well as a 3D model. Volume is calculated from this model. B) Similar steps were taken to measure CSF in the subarachnoid space. All measurements were done by one reader for consistency. C) T1-weighted MR images. Each row represents five coronal slices from one rat brain (top row = 3 month, second row = 12 month, third row = 20 month, bottom row = 30 month). Images were magnified and brightened for printing purposes. |
Intact relational memory and normal hippocampal structure in the early stage of psychosis
|
Publication: Biol Psychiatry. 2012 Jan 15;71(2):105-13. PMID: 22055016 | PDF Authors: Williams LE, Avery SN, Woolard AA, Heckers S. Institution: Department of Psychiatry, Vanderbilt University, Nashville, TN, USA. Background/Purpose: Previous studies indicate that the transition to psychosis is associated with dynamic changes of hippocampal integrity. Here we explored hippocampal volume and neural activation during a relational memory task in patients who were in the early stage of a psychotic illness. Forty-one early psychosis patients and 34 healthy control subjects completed a transitive inference (TI) task used previously in chronic schizophrenia patients. Participants learned to select the "winner" of two sets of stimulus pairs drawn from an overlapping sequence (A > B > C > D > E) and a nonoverlapping set (a > b, c > d, e > f, g > h). During a functional magnetic resonance imaging scan, participants were tested on the trained pairs and made inferential judgments on novel pairings that could be solved based on training (e.g., B vs. D). Hippocampal volumes were manually segmented and compared between groups. Functional magnetic resonance imaging analyses included 27 early psychosis patients and 30 control subjects who met memory training criteria. Groups did not differ on inference performance or hippocampal volume and exhibited similar activation of medial temporal regions when judging nonoverlapping pairs. However, patients who failed to meet memory training criteria had smaller hippocampal volumes. Neural activity during TI was less widespread in early psychosis patients, but between-group differences were not significant. Hippocampal activity during TI was positively correlated with inference performance only in control subjects. Our results provide evidence that relational memory impairment and hippocampal abnormalities, well established in chronic schizophrenia, are not fully present in early psychosis patients. This provides a rationale for early intervention, targeting the possible delay, reduction, or prevention of these deficits. Funding:
|
 Significant clusters of activation during transitive inference, p < .05 corrected cluster threshold: bilateral medial and superior frontal gyri (A control), bilateral cingulate (B, C, D controls, C early psychosis), middle temporal gyrus (left B, C, D control, bilateral D, E early psychosis), left superior temporal gyrus (B control), left paracentral lobule (B control), bilateral angular gyrus (D control), bilateral precuneus (D early psychosis), bilateral middle/inferior occipital gyrus (E control), and bilateral cerebellum (D, E). A direct comparison between groups revealed no significant differences. |
TREK: an integrated system architecture for intraoperative cone-beam CT-guided surgery
|
Publication: Int J Comput Assist Radiol Surg. 2012 Jan;7(1):159-73. PMID: 21744085 Authors: Uneri A, Schafer S, Mirota DJ, Nithiananthan S, Otake Y, Taylor RH, Siewerdsen JH. Institution: Department of Computer Science, Johns Hopkins University, Baltimore, MD, USA. Background/Purpose: A system architecture has been developed for integration of intraoperative 3D imaging [viz., mobile C-arm cone-beam CT (CBCT)] with surgical navigation (e.g., trackers, endoscopy, and preoperative image and planning data). The goal of this paper is to describe the architecture and its handling of a broad variety of data sources in modular tool development for streamlined use of CBCT guidance in application-specific surgical scenarios. METHODS: The architecture builds on two proven open-source software packages, namely the cisst package (Johns Hopkins University, Baltimore, MD) and 3D Slicer (Brigham and Women's Hospital, Boston, MA), and combines data sources common to image-guided procedures with intraoperative 3D imaging. Integration at the software component level is achieved through language bindings to a scripting language (Python) and an object-oriented approach to abstract and simplify the use of devices with varying characteristics. The platform aims to minimize offline data processing and to expose quantitative tools that analyze and communicate factors of geometric precision online. Modular tools are defined to accomplish specific surgical tasks, demonstrated in three clinical scenarios (temporal bone, skull base, and spine surgery) that involve a progressively increased level of complexity in toolset requirements. The resulting architecture (referred to as "TREK") hosts a collection of modules developed according to application-specific surgical tasks, emphasizing streamlined integration with intraoperative CBCT. These include multi-modality image display; 3D-3D rigid and deformable registration to bring preoperative image and planning data to the most up-to-date CBCT; 3D-2D registration of planning and image data to real-time fluoroscopy; infrared, electromagnetic, and video-based trackers used individually or in hybrid arrangements; augmented overlay of image and planning data in endoscopic or in-room video; and real-time "virtual fluoroscopy" computed from GPU-accelerated digitally reconstructed radiographs (DRRs). Application in three preclinical scenarios (temporal bone, skull base, and spine surgery) demonstrates the utility of the modular, task-specific approach in progressively complex tasks. The design and development of a system architecture for image-guided surgery has been reported, demonstrating enhanced utilization of intraoperative CBCT in surgical applications with vastly different requirements. The system integrates C-arm CBCT with a broad variety of data sources in a modular fashion that streamlines the interface to application-specific tools, accommodates distinct workflow scenarios, and accelerates testing and translation of novel toolsets to clinical use. The modular architecture was shown to adapt to and satisfy the requirements of distinct surgical scenarios from a common code-base, leveraging software components arising from over a decade of effort within the imaging and computer-assisted interventions community. Funding:
|
2011
Atlas-guided segmentation of vervet monkey brain MRI
|
Publication: Open Neuroimag J. 2011 Dec;5:186-97. PMID: 22253661 | PDF Authors: Fedorov A, Li X, Pohl KM, Bouix S, Styner M, Addicott M, Wyatt C, Daunais JB, Wells WM, Kikinis R. Institution: Surgical Planning Laboratory, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA. Background/Purpose: User interaction is required for reliable segmentation of brain tumors in clinical practice and in clinical research. By incorporating current research tools, 3D Slicer provides a set of interactive, easy to use tools that can be efficiently used for this purpose. One of the modules of 3D Slicer is an interactive editor tool, which contains a variety of interactive segmentation effects. Use of these effects for fast and reproducible segmentation of a single glioblastoma from magnetic resonance imaging data is demonstrated. The innovation in this work lies not in the algorithm, but in the accessibility of the algorithm because of its integration into a software platform that is practical for research in a clinical setting. Funding:
|
|
Publication: IEEE Transactions on Medical Imaging 2011 Dec; Authors: Daniel J. Mirota, Hanzi Wang, Russell H. Taylor, Masaru Ishii, Gary L. Gallia, Gregory D. Hager Institution: Department of Computer Science, Johns Hopkins University, Baltimore, MD, USA. Background/Purpose: Surgeries of the skull base require accuracy to safely navigate the critical anatomy. This is particularly the case for endoscopic endonasal skull base surgery (ESBS) where the surgeons work within millimeters of neurovascular structures at the skull base. Today’s navigation systems provide approximately 2 mm accuracy. Accuracy is limited by the indirect relationship of the navigation system, the image and the patient. We propose a method to directly track the position of the endoscope using video data acquired from the endoscope camera. Our method first tracks image feature points in the video and reconstructs the image feature points to produce three-dimensional (3D) points, and then registers the reconstructed point cloud to a surface segmented from pre-operative computed tomography (CT) data. After the initial registration, the system tracks image features and maintains the two-dimensional (2D)-3D correspondence of imagefeatures and 3D locations. These data are then used to update the current camera pose. We present a method for validation of our system, which achieves sub-millimeter (0.70 mm mean) target registration error (TRE) results. |
Comparison of Acute and Chronic Traumatic Brain Injury using Semi-automatic Multimodal Segmentation of MR Volumes
|
Publication: J Neurotrauma. 2011 Nov;28(11):2287-306. PMID: 21787171 | PDF Authors: Irimia A, Chambers MC, Alger JR, Filippou M, Prastawa MW, Wang B, Hovda DA, Gerig G, Toga AW, Kikinis R, Vespa PM, Van Horn JD. Institution: Laboratory of Neuro Imaging, University of California, Los Angeles, California 90095, USA. Background/Purpose: Though neuroimaging is essential for prompt and proper management of TBI, there is regrettable and acute lack of robust methods for the visualization and assessment of TBI pathophysiology, especially for of the purpose of improving clinical outcome metrics. Until now, the application of automatic segmentation algorithms to TBI in a clinical setting has remained an elusive goal because existing methods have, for the most part, been insufficiently robust to faithfully capture TBI-related changes in brain anatomy. This article introduces and illustrates the combined use of multimodal TBI segmentation and time point comparison using 3D Slicer, a widely-used software environment whose TBI data processing solutions are openly available. For three representative TBI cases, semi-automatic tissue classification and 3D model generation are performed to perform intra-patient time point comparison of TBI using multimodal volumetrics and clinical atrophy measures. Identification and quantitative assessment of extra- and intra-cortical bleeding, lesions, edema and diffuse axonal injury are demonstrated. The proposed tools allow cross-correlation of multimodal metrics from structural imaging (structural volume, atrophy measurements, etc.) with clinical outcome variables and other potential factors predictive of recovery. In addition, the workflows described are suitable for TBI clinical practice and patient monitoring, particularly for assessing damage extent and for the measurement of neuroanatomical change over time. With knowledge of general location, extent, and degree of change, such metrics can be associated with clinical measures and subsequently used to suggest viable treatment options. Funding:
|
 Three-dimensional models of TBI anatomy for Patient 1 (acute baseline time point), as generated in 3D Slicer. Each row displays one of six canonical views of the brain, whereas each column corresponds to a structure type (ventricles, non-hemorrhagic lesions, hemorrhagic lesions, and full model). In (A), ventricles are shown as opaque, whereas extracerebral injuries, gray matter (GM) and white matter (WM) volumes are shown as transparent to facilitate visualization of each structure in relationship to cortical landmarks. In (B–D), ventricles are also shown as transparent for similar reasons. Arrows indicate TBI-related pathology of interest (see text for elaboration). |
3D Slicer
|
Publication: PDF Video. 2011 November 18. Authors: John Muschelli, Vadim Zipunnikov Institution: Department of Biostatistics, Johns Hopkins Bloomberg School of Public Health. Background/Purpose: Quick tutorial of how to make 3D models in 3D Slicer. |
An Open Environment CT-US Fusion for Tissue Segmentation during Interventional Guidance
|
Publication: PLoS One. 2011 Novemeber;6(11):e27372. PMID: 22132098 | PDF Authors: Caskey CF, Hlawitschka M, Qin S, Mahakian LM, Cardiff RD, Boone JM, Ferrara KW. Institution: Department of Biomedical Engineering, University of California Davis, Davis, CA, USA. Background/Purpose: Therapeutic ultrasound (US) can be noninvasively focused to activate drugs, ablate tumors and deliver drugs beyond the blood brain barrier. However, well-controlled guidance of US therapy requires fusion with a navigational modality, such as magnetic resonance imaging (MRI) or X-ray computed tomography (CT). Here, we developed and validated tissue characterization using a fusion between US and CT. The performance of the CT/US fusion was quantified by the calibration error, target registration error and fiducial registration error. Met-1 tumors in the fat pads of 12 female FVB mice provided a model of developing breast cancer with which to evaluate CT-based tissue segmentation. Hounsfield units (HU) within the tumor and surrounding fat pad were quantified, validated with histology and segmented for parametric analysis (fat: −300 to 0 HU, protein-rich: 1 to 300 HU, and bone: HU>300). Our open source CT/US fusion system differentiated soft tissue, bone and fat with a spatial accuracy of ~1 mm. Region of interest (ROI) analysis of the tumor and surrounding fat pad using a 1 mm2 ROI resulted in mean HU of 68±44 within the tumor and −97±52 within the fat pad adjacent to the tumor (p<0.005). The tumor area measured by CT and histology was correlated (r2 = 0.92), while the area designated as fat decreased with increasing tumor size (r2 = 0.51). Analysis of CT and histology images of the tumor and surrounding fat pad revealed an average percentage of fat of 65.3% vs. 75.2%, 36.5% vs. 48.4%, and 31.6% vs. 38.5% for tumors <75 mm3, 75–150 mm3 and >150 mm3, respectively. Further, CT mapped bone-soft tissue interfaces near the acoustic beam during real-time imaging. Combined CT/US is a feasible method for guiding interventions by tracking the acoustic focus within a pre-acquired CT image volume and characterizing tissues proximal to and surrounding the acoustic focus. Funding:
|
 Combined CT/US characterizes tissue. (a) CT slice of a hindlimb tumor corresponding to the US slice in (b). The segmented CT slice is shown in (c) and overlaid on the US image in (d). The combined segmented CT/US image identifies fat proximal to and surrounding the tumor during ultrasonic imaging, with registration accuracy on the order of 1 mm. Nearby bone is obvious in the CT image (c) but not immediately visualized by US (b). |
|
Publication: Brain Behav. 2011 November; 1(2): 125–134. PMID: 22399092 | PDF Authors: Cherbuin N, Sachdev PS, Anstey KJ. Institution: Centre for Mental Health Research, Australian National University, Canberra, Australia. Background/Purpose: Handedness has been found to be associated with structural and functional cerebral differences. Left handedness and mixed handedness also appear to be associated with an elevated risk of some developmental and immunological disorders that may contribute to pathological processes developing in ageing. Inconsistent reports show that left handedness may be more prevalent in early-onset as well as late-onset Alzheimer's disease, but might also be associated with slower decline. Such inconsistencies may be due to handedness being usually modeled as a binary construct while substantial evidence suggests it to be a continuous trait. The aim of this study was to investigate the relationship between brain structures known to be implicated in pathological ageing and strength and direction of handedness. The association between handedness and hippocampal and amygdalar atrophy was investigated in 327 cognitively healthy older individuals. Handedness was measured with the Edinburgh Inventory. Two measures were computed from this index, one reflecting the direction (left = 0/right = 1) and the other the degree of handedness (ranging from 0 to 1). Hippocampal and amygdalar volumes were manually traced on scans acquired 4 years apart. Regression analyses were used to assess the relationship between strength and direction of handedness and incident hippocampal and amygdalar atrophy. Analyses showed that strength but not direction of handedness was a significant predictor of hippocampal (Left: beta = 0.118, P = 0.013; Right: beta = 0.116, P = 0.010) and amygdalar (Right: beta = 0.105, P = 0.040) atrophy. The present findings suggest that mixed but not left handedness is associated with greater hippocampal and amygdalar atrophy. This effect may be due to genetic, environmental, or behavioural differences that will need further investigation in future studies. Fundings:
|
Multiple Costal Cartilage Graft Reconstruction for the Treatment of a Full-length Laryngotracheal Stenosis after an Inhalation Burn
|
Publication: Interact Cardiovasc Thorac Surg. 2011 Oct;13(4):453-5. PMID: 21798888 | PDF Authors: Furák J., Szakács L., Nagy A., Rovó L. Institution: Department of Oto-Rhino-Laryngology, Head and Neck Surgery, University of Szeged, Szeged, Hungary. Background/Purpose: After suffering an inhalation burn, a 22-year-old male was intubated for seven days. Full-length massive scar formation in the upper airway necessitated tracheostomy five months later. After this, the stenosis became complete in the cricoid region, and a long cannula was needed to maintain the severely damaged middle-distal trachea. After unsuccessful laser dilatation, the more stenotic 3 cm distal tracheal segment was resected, but two months later the stenosis recurred. As resection was ineffective, tracheoplasty was performed via a right-sided thoracotomy; the re-stenotized trachea was incised in length and successfully extended with 5 cm long, oval-shaped rib cartilage. Three months later, the complete cricotracheal stenosis was fixed by combined laryngofissure and cricoid laminotomy with two 6 cm×2.5 cm cartilage pieces sutured into the incisions. The middle portion of the trachea was expanded with a similar graft inserted into the anterior wall below the tracheostomy. The fixing T-tube was removed three months later, and the patient had an adequate airway two years after the last procedure. We conclude that multiple cartilage graft reconstruction can be successful even after the development of an extremely long airway stenosis following inhalation burn injury. |
A supervised patch-based approach for human brain labeling
|
Publication: IEEE Trans Med Imaging. 2011 Oct;30(10):1852-62. PMID: 21606021 | PDF Authors: Rousseau F, Habas PA, Studholme C. Institution: Laboratoire des Sciences de l’Image, de l’Informatique et de la Télédétection (LSIIT), University of Strasbourg, Illkirch, France. Background/Purpose: We propose in this work a patch-based image labeling method relying on a label propagation framework. Based on image intensity similarities between the input image and an anatomy textbook, an original strategy which does not require any nonrigid registration is presented. Following recent developments in nonlocal image denoising, the similarity between images is represented by a weighted graph computed from an intensity-based distance between patches. Experiments on simulated and in vivo magnetic resonance images show that the proposed method is very successful in providing automated human brain labeling. Funding:
|
The chronic bronchitic phenotype of COPD: an analysis of the COPDGene Study
|
Publication: Chest. 2011 Sep;140(3):626-33. PMID: 21474571 | PDF Authors: Kim V, Han MK, Vance GB, Make BJ, Newell JD, Hokanson JE, Hersh CP, Stinson D, Silverman EK, Criner GJ; COPDGene Investigators. Institution: Temple University School of Medicine, Philadelphia, PA, USA. Background/Purpose: Chronic bronchitis (CB) in patients with COPD is associated with an accelerated lung function decline and an increased risk of respiratory infections. Despite its clinical significance, the chronic bronchitic phenotype in COPD remains poorly defined. We analyzed data from subjects enrolled in the Genetic Epidemiology of COPD (COPDGene) Study. A total of 1,061 subjects with GOLD (Global Initiative for Chronic Obstructive Lung Disease) stage II to IV were divided into two groups: CB (CB+) if subjects noted chronic cough and phlegm production for ≥ 3 mo/y for 2 consecutive years, and no CB (CB-) if they did not. There were 290 and 771 subjects in the CB+ and CB- groups, respectively. Despite similar lung function, the CB+ group was younger (62.8 ± 8.4 vs 64.6 ± 8.4 years, P = .002), smoked more (57 ± 30 vs 52 ± 25 pack-years, P = .006), and had more current smokers (48% vs 27%, P < .0001). A greater percentage of the CB+ group reported nasal and ocular symptoms, wheezing, and nocturnal awakenings secondary to cough and dyspnea. History of exacerbations was higher in the CB+ group (1.21 ± 1.62 vs 0.63 ± 1.12 per patient, P < .027), and more patients in the CB+ group reported a history of severe exacerbations (26.6% vs 20.0%, P = .024). There was no difference in percent emphysema or percent gas trapping, but the CB+ group had a higher mean percent segmental airway wall area (63.2% ± 2.9% vs 62.6% ± 3.1%, P = .013). CB in patients with COPD is associated with worse respiratory symptoms and higher risk of exacerbations. This group may need more directed therapy targeting chronic mucus production and smoking cessation not only to improve symptoms but also to reduce risk, improve quality of life, and improve outcomes. Funding:
|
Airway Inspector: Phenotyping the Lung in COPD
|
Publication: Airway Inspector Contact Person: Raul San Jose Estepar Institution: Surgical Planning Laboratory at Brigham and Women's Hospital, Boston, MA. Background/Purpose: Airway Inspector is a free open source tool for CT-based image quantitative analysis of the lung. It provides the tools that clinical scientist can used to explore airway morphology and desitometric characteristics. Airway Inspector is the gateway to phenotype airway diseases. Airway Inspector is maintained by the Applied Chest Imaging Laboratory (ACIL) and is based on the 3D Slicer. The 3D Slicer is a general purposed open source platform for medical image analysis maintained by the Surgical Planning Laboratory at Brigham and Women's Hospital, Boston, MA. Funding:
|
Distance Measurement in Middle Ear Surgery using a Telemanipulator
|
Publication: Med Image Comput Comput Assist Interv. 2011;14(Pt 1):41-8. PMID: 22003598 | PDF Authors: Maier T, Strauss G, Bauer F, Grasser A, Hata N, Lueth TC. Institution: Institute of Microtechnology and Medical Device Technology (MiMed), Technische Universität München, Germany. Background/Purpose: In this article, a new tool for the intraoperative measurement of distances within the middle ear by means of a micromanipulator is presented. The purpose of this work was to offer the surgeon a highly accurate tool for measuring the distances between two points in the 3D operational field. The tool can be useful in various operations; this article focuses, however, on measuring the distance between the stapes footplate and the long process of the incus of the middle ear. This distance is important for estimating the proper prosthesis length in stapedotomy for treating otosclerosis. We evaluated the system using a simplified mechanical model. Our results show that the system can measure distances with a maximum error of 0.04 mm.
|
3D Slicer as a tool for interactive brain tumor segmentation
|
Publication: Conf Proc IEEE Eng Med Biol Soc. 2011 Aug;2011:6982-4. PMID: 22255945 Authors: Kikinis R, Pieper S. Institution: Surgical Planning Laboratory, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA. Background/Purpose: User interaction is required for reliable segmentation of brain tumors in clinical practice and in clinical research. By incorporating current research tools, 3D Slicer provides a set of interactive, easy to use tools that can be efficiently used for this purpose. One of the modules of 3D Slicer is an interactive editor tool, which contains a variety of interactive segmentation effects. Use of these effects for fast and reproducible segmentation of a single glioblastoma from magnetic resonance imaging data is demonstrated. The innovation in this work lies not in the algorithm, but in the accessibility of the algorithm because of its integration into a software platform that is practical for research in a clinical setting. Funding:
|
The Effect of Augmented Reality Training on Percutaneous Needle Placement in Spinal Facet Joint Injections
|
Publication: IEEE Trans Biomed Eng. 2011 Jul;58(7):2031-7. PMID: 21435970 | PDF Authors: Caitlin T. Yeo, Tamas Ungi, Paweena U-Thainual, Andras Lasso, Robert C. McGraw, Gabor Fichtinger Institution: School of Medicine, Queen’s University, Kingston ON K7L 3N6, Canada Background/Purpose: The purpose of this study was to determine if augmented reality image overlay and laser guidance systems can assist learn how to perform the procedure without the risk of errors medical trainees in learning the correct placement of a needle for causing harm to the patient [2]. Additionally, simulation allows percutaneous facet joint injection. The Perk Station training suite was used to conduct and record the needle insertion procedures. A total of 40 volunteers were randomized into two groups of 20. 1) The Overlay group received a training session that consisted of four insertions with image and laser guidance, followed by two insertions with laser overlay only. 2) The Control group received a training session of six classical freehand insertions. Both groups then conducted two freehand insertions. The movement of the needle was tracked during the series of insertions. The final insertion procedure was assessed to determine if there was a benefit to the overlay method compared to the freehand insertions. The Overlay group had a better success rate (83.3% versus 68.4%, p = 0.002), and potential for less tissue damage as measured by the amount of needle movement inside the phantom (3077.6 mm2 versus 5607.9 mm2, p = 0.01). These results suggest that an augmented reality overlay guidance system can assist medical trainees in acquiring technical competence in a percutaneous needle insertion procedure. Funding:
|
Optimal transseptal puncture location for robot-assisted left atrial catheter ablation
|
Publication: Int J Med Robot. 2011 Jun;7(2):193-201. PMID: 21538767 | PDF Authors: Jagadeesan Jayender, Rajni V. Patel, Gregory F. Michaud, Nobuhiko Hata Institution: Surgical Planning Laboratory, Department of Radiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA. Background/Purpose: The preferred method of treatment for atrial fibrillation (AF) is by catheter ablation, in which a catheter is guided into the left atrium through a transseptal puncture. However, the transseptal puncture constrains the catheter, thereby limiting its manoeuvrability and increasing the difficulty in reaching various locations in the left atrium. In this paper, we address the problem of choosing the optimal transseptal puncture location for performing cardiac ablation to obtain maximum manoeuvrability of the catheter. We have employed an optimization algorithm to maximize the global isotropy index (GII) to evaluate the optimal transseptal puncture location. As part of this algorithm, a novel kinematic model for the catheter has been developed, based on a continuum robot model. Pre-operative MR/CT images of the heart are segmented using the open source image-guided therapy software, 3D Slicer, to obtain models of the left atrium and septal wall. These models are input to the optimization algorithm to evaluate the optimal transseptal puncture location. Funding:
|
Multiple indices of diffusion identifies white matter damage in mild cognitive impairment and Alzheimer's disease
|
Publication: PLoS One. 2011 June;6(6):e21745. PMID: 21738785 | PDF Authors: O'Dwyer L, Lamberton F, Bokde AL, Ewers M, Faluyi YO, Tanner C, Mazoyer B, O'Neill D, Bartley M, Collins DR, Coughlan T, Prvulovic D, Hampel H. Institution: Department of Psychiatry, Psychosomatic Medicine and Psychotherapy, Goethe University, Frankfurt, Germany. Background/Purpose: The study of multiple indices of diffusion, including axial (DA), radial (DR) and mean diffusion (MD), as well as fractional anisotropy (FA), enables WM damage in Alzheimer's disease (AD) to be assessed in detail. Here, tract-based spatial statistics (TBSS) were performed on scans of 40 healthy elders, 19 non-amnestic MCI (MCIna) subjects, 14 amnestic MCI (MCIa) subjects and 9 AD patients. Significantly higher DA was found in MCIna subjects compared to healthy elders in the right posterior cingulum/precuneus. Significantly higher DA was also found in MCIa subjects compared to healthy elders in the left prefrontal cortex, particularly in the forceps minor and uncinate fasciculus. In the MCIa versus MCIna comparison, significantly higher DA was found in large areas of the left prefrontal cortex. For AD patients, the overlap of FA and DR changes and the overlap of FA and MD changes were seen in temporal, parietal and frontal lobes, as well as the corpus callosum and fornix. Analysis of differences between the AD versus MCIna, and AD versus MCIa contrasts, highlighted regions that are increasingly compromised in more severe disease stages. Microstructural damage independent of gross tissue loss was widespread in later disease stages. Our findings suggest a scheme where WM damage begins in the core memory network of the temporal lobe, cingulum and prefrontal regions, and spreads beyond these regions in later stages. DA and MD indices were most sensitive at detecting early changes in MCIa. |
 Analysis of disease progression with MCIna versus AD, and, MCIa versus AD. MCIna versus AD is shown in red, MCIa versus AD is shown in green, and areas where these contrasts overlap is shown in blue. (a) FA decreases together with DR increases. (b) FA decreases together with MD increases. (c) FA decreases where MD remains unchanged. See text main text for anatomical description and explanation of the three combinations of indices. Areas of significant difference were visualized using the 3D Slicer program (http://www.slicer.org/). |
Assessment of abdominal adipose tissue and organ fat content by magnetic resonance imaging
|
Publication: Obes Rev. 2011 May;12(5):e504-15. PMID: 21348916 | PDF Authors: Hu HH, Nayak KS, Goran MI. Institution: Ming Hsieh Department of Electrical Engineering, Viterbi School of Engineering, University of Southern California, Los Angeles, CA, USA. Background/Purpose: As the prevalence of obesity continues to rise, rapid and accurate tools for assessing abdominal body and organ fat quantity and distribution are critically needed to assist researchers investigating therapeutic and preventive measures against obesity and its comorbidities. Magnetic resonance imaging (MRI) is the most promising modality to address such need. It is non-invasive, utilizes no ionizing radiation, provides unmatched 3-D visualization, is repeatable, and is applicable to subject cohorts of all ages. This article is aimed to provide the reader with an overview of current and state-of-the-art techniques in MRI and associated image analysis methods for fat quantification. The principles underlying traditional approaches such as T(1) -weighted imaging and magnetic resonance spectroscopy as well as more modern chemical-shift imaging techniques are discussed and compared. The benefits of contiguous 3D acquisitions over 2D multislice approaches are highlighted. Typical post-processing procedures for extracting adipose tissue depot volumes and percent organ fat content from abdominal MRI data sets are explained. Furthermore, the advantages and disadvantages of each MRI approach with respect to imaging parameters, spatial resolution, subject motion, scan time and appropriate fat quantitative endpoints are also provided. Practical considerations in implementing these methods are also presented. Funding:
|
 Abdominal fat fraction volume from IDEAL, illustrating the contiguous 3D nature of the data. The volume consists of more than 70 native axial slices and was obtained with five 15 second breath-holds (R: right, L: left, A: anterior, P: posterior, I: inferior, S: superior). Rendering created with 3D Slicer software. |
|
Publication: Neurobiol Aging. 2011 May;32(5):916-32. PMID: 19570593 | PDF Authors: Kristine B. Walhovd, Lars T. Westlye, Inge Amlien, Thomas Espeseth, Ivar Reinvang, Naftali Raz, Ingrid Agartz, David H. Salat, Doug N. Greve, Bruce Fischl, Anders M. Dale, Anders M. Fjell Institution: Center for the Study of Human Cognition, Department of Psychology, University of Oslo, Norway. Background/Purpose: Magnetic resonance imaging (MRI) is the principal method for studying structural age-related brain changes in vivo. However, previous research has yielded inconsistent results, precluding understanding of structural changes of the ageing brain. This inconsistency is due to methodological differences and/or different aging patterns across samples. To overcome these problems, we tested age effects on 17 different neuroanatomical structures and total brain volume across five samples, of which one was split to further investigate consistency (883 participants). Widespread age-related volume differences were seen consistently across samples. In four of the five samples, all structures, except the brainstem, showed age-related volume differences. The strongest and most consistent effects were found for cerebral cortex, pallidum, putamen and accumbens volume. Total brain volume, cerebral white matter, caudate, hippocampus and the ventricles consistently showed non-linear age functions. Healthy aging appears associated with more widespread and consistent age-related neuroanatomical volume differences than previously believed. Funding:
|
Brain Regional Lesion Burden and Impaired Mobility in the Elderly
|
Publication: Neurobiol Aging. 2011 Apr;32(4):646-54. PMID: 19428145 | PDF Authors: Moscufo N, Guttmann C, Meier D, Csapo I, Hildenbrand P, Healy B, Schmidt J, Wolfson L Institution: Center for Neurological Imaging, Department of Radiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA. Background/Purpose: This study investigated the relationship of brain white matter (WM) lesions affecting specific neural networks with decreased mobility in ninety-nine healthy community-dwelling subjects ≥75 years old prospectively enrolled by age and mobility status. We assessed lesion burden in the genu, body and splenium of corpus callosum; anterior, superior and posterior corona radiata; anterior and posterior limbs of internal capsule; corticospinal tract; and superior longitudinal fasciculus. Burden in the splenium of corpus callosum (SCC) demonstrated the highest correlation particularly with walking speed (r=0.4, p<10-4), and in logistic regression it was the best regional predictor of low mobility performance. We also found that independent of mobility, corona radiata has the largest lesion burden with anterior (ACR) and posterior (PCR) aspects being the most frequently affected. The results suggest that compromised inter-hemispheric integration of visuospatial information through the SCC plays an important role in mobility impairment in the elderly. The relatively high lesion susceptibility of ACR and PCR in all subjects may obscure the importance of these lesions in mobility impairment. 3D Slicer application has been used in all the imaging analysis steps, which included intracranial cavity outline, one- and two-channel brain tissue segmentation, labelmap outputs expert review and manual correction, image comparison, volume determination, and three-dimensional modeling. Funding:
|
 White matter lesion distribution and frequency (color bar) in the study subjects at baseline: normal (A), intermediate (B), and impaired (C) mobility. Panel D shows a 3D model reconstruction of the most affected structure, the corona radiata (green), within the white matter (transparent gray), and with ventricles (blue) also shown. |
Lung Volumes and Emphysema in Smokers with Interstitial Lung Abnormalities
|
Publication: N Engl J Med. 2011 Mar 10;364(10):897-906. PMID: 21388308 | PDF Authors: George R. Washko, Gary M. Hunninghake, Isis E. Fernandez, Yuka Okajima, Tsuneo Yamashiro, James C. Ross, Raúl San José Estépar, David A. Lynch, John M. Brehm, Katherine P. Andriole, Alejandro A. Diaz, Ramin Khorasani, Katherine D’Aco, Frank C. Sciurba, Edwin K. Silverman, Hiroto Hatabu, Ivan O. Rosas Institution: Pulmonary and Critical Care Division, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA Background/Purpose: Cigarette smoking is associated with emphysema and radiographic interstitial lung abnormalities. The degree to which interstitial lung abnormalities are associated with reduced total lung capacity and the extent of emphysema is not known. We looked for interstitial lung abnormalities in 2416 (96%) of 2508 high-resolution computed tomographic (HRCT) scans of the lung obtained from a cohort of smokers. We used linear and logistic regression to evaluate the associations between interstitial lung abnormalities and HRCT measurements of total lung capacity and emphysema. Quantitative measures of total lung capacity and emphysema were performed with the Airway Inspector, the tool based on 3D Slicer for lung image analysis. Funding:
|
 Four Major Radiographic Subtypes of Interstitial Lung Abnormalities. Each row of radiographs and reconstructions represents data from a single study participant. The images in Panel A are characteristic of centrilobular interstitial lung abnormalities; Panel B, subpleural interstitial lung abnormalities; Panel C, mixed centrilobular and subpleural interstitial lung abnormalities; and Panel D, radiographic interstitial lung disease. The images in the first two columns are three-dimensional reconstructions of an anterior-to-posterior view of the lungs (column 1) and a caudal-to-cephalad view (column 2). The translucent yellow–green represents the lung parenchyma; white, the tracheobronchial tree; opaque green, centrilobular opacities; and opaque orange, subpleural abnormalities. Columns 3 and 4 present axial high-resolution computed tomographic images of the chest, with images in column 3 approximately at the level of the carina and those in column 4 approximately at the level of the right inferior pulmonary vein. |
Integration of 3D anatomical data obtained by CT imaging and 3D optical scanning for computer aided implant surgery
|
Publication: BMC Medical Imaging 2011 Feb; 11:5. PMID: 21338504 | PDF Authors: Gianni Frisardi, Giacomo Chessa, Sandro Barone, Alessandro Paoli, Armando Razionale and Flavio Frisardi Institution: "Epochè" Orofacial Pain Center, Nettuno (Rome), Italy. Background/Purpose: A precise placement of dental implants is a crucial step to optimize both prosthetic aspects and functional constraints. In this context, the use of virtual guiding systems has been recognized as a fundamental tool to control the ideal implant position. In particular, complex periodontal surgeries can be performed using preoperative planning based on CT data. The critical point of the procedure relies on the lack of accuracy in transferring CT planning information to surgical field through custom-made stereo-lithographic surgical guides. Methods: In this work, a novel methodology is proposed for monitoring loss of accuracy in transferring CT dental information into periodontal surgical field. The methodology is based on integrating 3D data of anatomical (impression and cast) and preoperative (radiographic template) models, obtained by both CT and optical scanning processes. A clinical case, relative to a fully edentulous jaw patient, has been used as test case to assess the accuracy of the various steps concurring in manufacturing surgical guides. In particular, a surgical guide has been designed to place implants in the bone structure of the patient. The analysis of the results has allowed the clinician to monitor all the errors, which have been occurring step by step manufacturing the physical templates. The use of an optical scanner, which has a higher resolution and accuracy than CT scanning, has demonstrated to be a valid support to control the precision of the various physical models adopted and to point out possible error sources. A case study regarding a fully edentulous patient has confirmed the feasibility of the proposed methodology. |
Anisotropy of Transcallosal Motor Fibres Indicates Functional Impairment in Children with Periventricular Leukomalacia
|
Publication: Dev Med Child Neurol. 2011 Feb;53(2):179-86. PMID: 21121906 | PDF Authors: Koerte I, Pelavin P, Kirmess B, Fuchs T, Berweck S, Laubender RP, Borggraefe I, Schroeder S, Danek A, Rummeny C, Reiser M, Kubicki M, Shenton ME, Ertl-Wagner B, Heinen F. Institution: Institute of Clinical Radiology, Ludwig-Maximilians-University, Munich, Germany. Background/Purpose: In children with bilateral spastic cerebral palsy (CP), periventricular leukomalacia (PVL) is commonly identified on magnetic resonance imaging. We characterized this white matter condition by examining callosal microstructure, interhemispheric inhibitory competence (IIC), and mirror movements. Method We examined seven children (age range 11y 9mo-17y 9mo, median age 15y 10mo, four females, three males) with bilateral spastic CP/PVL (Gross Motor Function Classification System level I or II, Manual Ability Classification System level I) and 12 age-matched controls (age range 11y 7mo-17y 1mo, median age 15y 6mo, seven females, five males). Fractional anisotropy of the transcallosal motor fibres (TCMF) and the corticospinal tract (CST) of both sides were calculated. The parameters of IIC (transcranial magnetic stimulation) and mirror movements were measured using a standardized clinical examination and a computer-based hand motor test. Results Fractional anisotropy was lower in children with bilateral spastic CP/PVL regarding the TCMF, but not the left or right CST. Resting motor threshold was elevated in children with bilateral spastic CP/PVL whereas measures of IIC tended to be lower. Mirror movements were markedly elevated in bilateral spastic CP/PVL. Interpretation This study provides new information on different aspects of motor function in children with bilateral spastic CP/PVL. Decreased fractional anisotropy of TCMF is consistent with impairment of hand motor function in children with bilateral spastic CP/PVL. The previously overlooked microstructure of the TCMF may serve as a potential indicator for hand motor function in patients with bilateral spastic CP/PVL. Funding:
|
Automatic lung segmentation in CT images with accurate handling of the hilar region
|
Publication: J Digit Imaging. 2011 Feb;24(1):11-27. PMID: 19826872 | PDF Authors: De Nunzio G, Tommasi E, Agrusti A, Cataldo R, De Mitri I, Favetta M, Maglio S, Massafra A, Quarta M, Torsello M, Zecca I, Bellotti R, Tangaro S, Calvini P, Camarlinghi N, Falaschi F, Cerello P, Oliva P. Institution: Department of Materials Science, University of Salento, and INFN, Lecce, Italy. Background/Purpose: A fully automated and three-dimensional (3D) segmentation method for the identification of the pulmonary parenchyma in thorax X-ray computed tomography (CT) datasets is proposed. It is meant to be used as pre-processing step in the computer-assisted detection (CAD) system for malignant lung nodule detection that is being developed by the Medical Applications in a Grid Infrastructure Connection (MAGIC-5) Project. In this new approach the segmentation of the external airways (trachea and bronchi), is obtained by 3D region growing with wavefront simulation and suitable stop conditions, thus allowing an accurate handling of the hilar region, notoriously difficult to be segmented. Particular attention was also devoted to checking and solving the problem of the apparent 'fusion' between the lungs, caused by partial-volume effects, while 3D morphology operations ensure the accurate inclusion of all the nodules (internal, pleural, and vascular) in the segmented volume. The new algorithm was initially developed and tested on a dataset of 130 CT scans from the Italung-CT trial, and was then applied to the ANODE09-competition images (55 scans) and to the LIDC database (84 scans), giving very satisfactory results. In particular, the lung contour was adequately located in 96% of the CT scans, with incorrect segmentation of the external airways in the remaining cases. Segmentation metrics were calculated that quantitatively express the consistency between automatic and manual segmentations: the mean overlap degree of the segmentation masks is 0.96 ± 0.02, and the mean and the maximum distance between the mask borders (averaged on the whole dataset) are 0.74 ± 0.05 and 4.5 ± 1.5, respectively, which confirms that the automatic segmentations quite correctly reproduce the borders traced by the radiologist. Moreover, no tissue containing internal and pleural nodules was removed in the segmentation process, so that this method proved to be fit for the use in the framework of a CAD system. Finally, in the comparison with a two-dimensional segmentation procedure, inter-slice smoothness was calculated, showing that the masks created by the 3D algorithm are significantly smoother than those calculated by the 2D-only procedure. Fundings:
|
Using 3D Slicer in Astronomy
|
Institution: The Initiative in Innovative Computing (IIC), Harvard University, Cambridge, Massachusetts Project: IIC AstroMed Project Background/Purpose: While astronomy and medical imaging seem very different, both fields search large amounts of image data looking for meaningful patterns. For example, a physician may inspect a patient's MRI scans looking for signs of disease, while an astronomer will analyze radio telescope image data to find evidence of a new star being born. The two sciences have separately developed many techniques to analyze, visualize, and catalog complex multi-dimensional imaging data, but seldom have experts from the two areas worked together. The 3D Slicer software package is AstroMed's core tool for astronomical 3D data visualization. Originally designed for medical image analysis, 3D Slicer has many capabilities that are uncommon in astronomy software. The AstroMed project brings together researchers from the Harvard Medical School and the Harvard-Smithsonian Center for Astrophysics, along with both national and international collaborators, to combine their knowledge and advance the state-of-the-art in both medical imaging and astronomy. Related article in the February 11, 2011 issue of Science Magazine. |
Brain morphometry in autism spectrum disorders: a unified approach for structure-specific statistical analysis of neuroimaging data
|
Publication: Biomed Sci Instrum. 2011;47:135-41. PMID: 21525610 Authors: Vatta F, Di Salle F. Institution: Maastricht University, Maastricht, The Netherlands. Background/Purpose: Autism spectrum disorders (ASD) are a neurodevelopmental condition with multiple causes, comorbid conditions, and a wide range in the type and severity of symptoms expressed by different individuals. This makes the neuroanatomy of autism inherently difficult to describe. It has been assumed in the scientific literature that deviations in regional brain size in clinical samples are directly related to maldevelopment or pathogenesis. The performed clinical studies analyzed specific brain structures that are assumed to be correlated to autistic brain behaviors. Examples of performed analyses, based upon manual or semi-automated segmentation from magnetic resonance imaging (MRI) scans, include volumetric measures of specific brain structures, or small groups of structures, as caudate, corpus callosum, putamen, hippocampus, nucleus accumbens, evaluating differences between groups of subjects with autism and control subjects. Nonetheless, the brain regions analyzed that differ between patients and control subjects have not been always consistent over the performed studies. This inconsistency might be due to the fact that the specific single volume differences that have been reported in the literature for the different brain structures under investigation may, instead, be not independent during pathogenesis. Hence, this issue comes into play in logically framing a comprehensive assessment of putative abnormalities in regional brain volumes. To this aim, a whole brain investigation system for a semi-automated morphometric statistical analysis of brain anatomy is presented in this paper and validated on a selected group of patients diagnosed with ASD that completed a 1.5 T magnetic resonance image (MRI) of the brain. The proposed system, which is mainly built basing upon the FreeSurfer and the 3D Slicer software frameworks for the volumetric analysis of brain imaging data, lies its foundations on the higher statistical power of the region of interest (ROI) approach, but equally aims at a higher exploratory power as it doesn't restrict its focus to a small number of specific regions, thanks to a whole brain unified approach. |
Three dimensional in vivo modelling of vestibular schwannomas and surrounding cranial nerves using diffusion imaging tractography
|
Publication: Neurosurgery. 2011 Apr;68(4):1077-83. PMID: 21242825 Authors: Chen DQ, Quan J, Guha A, Tymianski M, Mikulis D, Hodaie M. Institution: Division of Neurosurgery, Toronto Western Hospital and University of Toronto, Toronto, Ontario, Canada. Background/Purpose: Preservation of cranial nerves (CN) is of paramount concern in the treatment of vestibular schwannomas (VS), particularly in large tumors with thinned and distorted CN fibers. However, imaging of the CN fibers surrounding VS has been limited using 2D imaging alone. We assessed whether tractography of the CN combined with anatomical MR imaging of the tumor can provide superior 3D visualization of tumor/CN complexes. 3T MR imaging including diffusion tensor imaging (DTI) and anatomical images were analyzed in three subjects with VS using 3D Slicer software. The DTI images were used to tract the courses of trigeminal, abducens, facial, and vestibulocochlear nerves. The anatomical images were used to model the 3D volume reconstruction of the tumor. The two sets of images were then superimposed using linear registration. Combined 3D tumor modelling and cranial nerve tractography can effectively and consistently reconstruct the 3D spatial relationship of CN/tumor complexes, and allows for superior visualization compared with 2D imaging. Lateral and superior distortion of the trigeminal nerve was observed in all cases. The position of the facial nerve was primarily anteriorly and inferiorly. The Gasserian ganglion and early postganglionic branches could also be visualized. Tractography and anatomical imaging were successfully combined to demonstrate the precise location of surrounding CN fibers. This technique can be useful in both neuronavigation and radiosurgical planning. Since knowledge of the course of these fibers is of important clinical interest, implementation of this technique may help decrease injury to cranial nerves during treatment of these lesions. |
|
Publication: Neurosurgery. 2011 Apr;68(4):1077-83. PMID: 21135719 Authors: Elhawary H, Liu H, Patel P, Norton I, Rigolo L, Papademetris X, Hata N, Golby AJ Institution: Surgical Planning Laboratory, Department of Radiology, Brigham and Women's Hospital, Harvard Medical School, 75 Francis Street, Boston, MA, 02115, USA. Background/Purpose: Brain surgery faces important challenges when trying to achieve maximum tumor resection while avoiding post-operative neurological deficits. In order for surgeons to have optimal intraoperative information concerning white matter anatomy, we developed a platform that allows the intra-operative real-time querying of tractography datasets during frameless stereotactic neuronavigation. Structural magnetic resonance imaging (MRI), functional MRI, and diffusion tensor imaging (DTI) were performed on 5 patients before undergoing lesion resection using neuronavigation. During the procedure, the tracked surgical tool tip position was transferred from the navigation system to the 3D Slicer software package, which used this position to seed the white matter tracts around the tool tip location, rendering a geometric visualization of these tracts on the pre-operative images previously loaded onto the navigation system. The clinical feasibility of this approach was evaluated during five cases of lesion resection. In addition, system performance was evaluated by measuring the latency between surgical tool tracking and visualization of the seeded white matter tracts. Lesion resection was performed successfully in all five patients. The seeded white matter tracts close to the lesion and other critical structures, as defined by the functional and structural images, were interactively visualized during the intervention to determine their spatial relationships relative to the lesion and critical cortical areas. Latency between tracking and visualization of tracts was less than a second for fiducial radius size of 4-5mm. Interactive tractography can provide an intuitive way of inspecting critical white matter tracts in the vicinity of the surgical region, allowing the surgeon to have increased intra-operative white matter information to execute the planned surgical resection. Funding:
|
 Images shown in the 3D Slicer using the intraoperative tractography navigation software module. The light green volume delineates the tumor location, the orange sphere represents the seed fiducial that is located at the tracked surgical tool tip location, the yellow regions indicate eloquent cortex as shown by language task-associated functional magnetic resonance imaging, and the tubular fibers represent the white matter tracts seeded by the fiducial location. White matter tracts are color coded according to their fractional anisotropy (color scale). A, white matter fibers being seeded by a fiducial representing tool tip position. Some infiltrating fibers can be seen within the segmented tumor volume (arrows). B, intraoperative querying of tractography data sets using the tool offset function. Fiducials F1 and F2 represent the surgical tool tip location and a fixed geometric offset respectively. The navigation module allows for geometric offsets from 280 to 80 mm from the tool tip position. The 2 fiducials can seed the tractography data set simultaneously. |
2010
Application of Vascular Model Toolkit (VMTK) for Coronary Arteries
|
Publication: Presented at 62nd Cardiological Society of India Conference, Kolkata, India, 2010 Dec; | PDF Authors: Christopher, J., Mohan Rao MB.,Chary, Duraikannu, Kishore,LT., Krishnam Raju Institution: Care Hospitals, Hyderabad, India Background/Purpose: Functional evaluation of coronary arteries involves study of their 3D shapes and diameters. Segmentation of vascular structures from 3D-Computerized Tomographic Angiogram (CTA) images can be achieved by using VMTK. This gives: (1) a polygonal surface model of vascular structures having centerline based geometric quantities and (2) compute centerlines and Maximal Inscribed Sphere Radius (MISR) of branching tubular structures (Voronoi image). VMTK module integrated in Slicer3 is used as it allows use of other Slicer3 modules for processing of images. We use: VesselEnhancement, EasyLevelsetSegmentation and Centerlines modules for study. |
Ion-Abrasion Scanning Electron Microscopy Reveals Surface-Connected Tubular Conduits in HIV-Infected Macrophages
|
Publication: PLoS Pathog. 2010 Sept;5(9):e1000591. PMID: 19779568 | PDF Authors: Bennett AE, Narayan K, Shi D, Hartnell LM, Gousset K, He H, Lowekamp BC, Yoo TS, Bliss D, Freed EO, Subramaniam S. Institution: Laboratory of Cell Biology, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA. Background/Purpose: HIV-1-containing internal compartments are readily detected in images of thin sections from infected cells using conventional transmission electron microscopy, but the origin, connectivity, and 3D distribution of these compartments has remained controversial. Here, we report the 3D distribution of viruses in HIV-1-infected primary human macrophages using cryo-electron tomography and ion-abrasion scanning electron microscopy (IA-SEM), a recently developed approach for nanoscale 3D imaging of whole cells. Using IA-SEM, we show the presence of an extensive network of HIV-1-containing tubular compartments in infected macrophages, with diameters of 150–200 nm, and lengths of up to 5 mm that extend to the cell surface from vesicular compartments that contain assembling HIV-1 virions. These types of surface-connected tubular compartments are not observed in T cells infected with the 29/31 KE Gag-matrix mutant where the virus is targeted to multi-vesicular bodies and released into the extracellular medium. IA-SEM imaging also allows visualization of large sheet-like structures that extend outward from the surfaces of macrophages, which may bend and fold back to allow continual creation of viral compartments and virion-lined channels. This potential mechanism for efficient virus trafficking between the cell surface and interior may represent a subversion of pre-existing vesicular machinery for antigen capture, processing, sequestration, and presentation. |
 3D representation of the surface and interior of an HIV-infected macrophage (animation is presented in Video S10). (A) Sections that would appear to contain ‘‘filopodia’’ when imaged by transmission electron microscopy of individual sections can actually correspond to large wavelike membrane processes as in this example. The virions are shown in red. (B, C) Schematic side (B) and front (C) views of these surface protrusions shown to indicate how bending and folding back of the extensions onto the surface of the cell could trap the contents of the aqueous environment within the invaginated folds of the membrane, and allow creation of viral compartments. |
MRI-Guided Robotic Prostate Biopsy: A Clinical Accuracy Validation
|
Publication: Int Conf Med Image Comput Comput Assist Interv. 2010 Sept;13(Pt 3):383-391. PMID: 20879423 | PDF Authors: Helen Xu, Andras Lasso, Siddharth Vikal, Peter Guion, Axel Krieger, Aradhana Kaushal, Louis L. Whitcomb, and Gabor Fichtinger Institutions: Queen's University, Kingston, Canada; National Institutes of Health, Bethesda, USA; Sentinelle Medical Inc., Toronto, Canada; Johns Hopkins University, Baltimore, USA Background/Purpose: Prostate cancer is a major health threat for men. For over five years, the U.S. National Cancer Institute has performed prostate biopsies with a magnetic resonance imaging (MRI)-guided robotic system. A retrospective evaluation methodology and analysis of the clinical accuracy of this system is reported. Using the pre and post-needle insertion image volumes, a registration algorithm that contains a two-step rigid registration followed by a deformable refinement was developed to capture prostate dislocation during the procedure. 3D Slicer was used to validate the registration algorithm by manually aligning the surfaces of segmented objects in 3D. Funding:
|
A method for planning safe trajectories in image-guided keyhole neurosurgery
|
Publication: Med Image Comput Comput Assist Interv. 2010 Sept;13(Pt 3):457-64. PMID: 20879432 | PDF Authors: Shamir RR, Tamir I, Dabool E, Joskowicz L, Shoshan Y. Institution: School of Engineering and Computer Science, The Hebrew University, Jerusalem, Israel. Background/Purpose: We present a new preoperative planning method for reducing the risk associated with insertion of straight tools in image-guided keyhole neurosurgery. The method quantifies the risks of multiple candidate trajectories and presents them on the outer head surface to assist the neurosurgeon in selecting the safest path. The surgeon can then define and/or revise the trajectory, add a new one using interactive 3D visualization, and obtain a quantitative risk measures. The trajectory risk is evaluated based on the tool placement uncertainty, on the proximity of critical brain structures, and on a predefined table of quantitative geometric risk measures. Our results on five targets show a significant reduction in trajectory risk and a shortening of the preoperative planning time as compared to the current routine method. Funding:
|
JHU Workshop for Talented Youth
|
Publication: Workshop for Talented Youth, 2010 Sept; Author: Christopher Wyatt, Ph.D. Institution: John Hopkins University, Baltimore, MD. Background/Purpose: This 2010 workshop for the JHU Center for Talented Youth was hosted at Virginia Tech and focused on neuroscience. The event included an interactive session titled "Brain Shape: Using images to predict the development of Alzheimer's Disease." The session included a short introduction to neuroimaging, followed by students, alone or in pairs, using the 3D Slicer software to compare healthy and pathological images of the brain. 100 students ranging in age from 13 to 16 years old attended the event. |
Canine Hippocampal Formation Composited into Three-dimensional Structure Using MPRAGE
|
Publication: J Vet Med Sci. 2010 Jul;72(7):853-60. PMID: 20179383 | PDF Authors: Jung MA, Lee MS, Lee IH, Lee AR, Jang DP, Kim YB, Cho ZH, Nahm SS, Eom KD Institution: Department of Veterinary Radiology and Diagnostic Imaging, College of Veterinary Medicine, Konkuk University. Background/Purpose: This study was performed to anatomically illustrate the living canine hippocampal formation in three-dimensions (3D), and to evaluate its relationship to surrounding brain structures. Three normal beagle dogs were scanned on a MR scanner with inversion recovery segmented three- dimensoinal gradient echo sequence (known as MP-RAGE: Magnetization Prepared Rapid Gradient Echo). The MRI data was manually segmented and reconstructed into a three-dimensional model using the 3D Slicer software tool. From the three-dimensional model, the spatial relationships between hippocampal formation and surrounding structures were evaluated. With the increased spatial resolution and contrast of the MPRAGE, the canine hippocampal formation was easily depicted. The reconstructed three-dimensional image allows easy understanding of the hippocampal contour and demonstrates the structural relationship of the hippocampal formation to surrounding structures in vivo. Funding:
|
|
Publication: J Am Coll Surg. 2010 Jun;210(6):927-33. PMID: 20510801 | PDF Authors: Tomikawa M, Hong J, Shiotani S, Tokunaga E, Konishi K, Ieiri S, Tanoue K, Akahoshi T, Maehara Y, Hashizume M. Institution: Department of Future Medicine and Innovative Medical Information, Graduate School of Medical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka, Japan. Background/Purpose: The aim of this study was to report on the early experiences using a real-time 3-dimensional (3D) virtual reality navigation system with open MRI for breast-conserving surgery. We developed a real-time 3D virtual reality navigation system with open MRI, and evaluated the mismatch between the navigation system and real distance using a 3D phantom. Two patients with nonpalpable MRI-detected breast tumors underwent breast-conserving surgery under the guidance of the navigation system. An initial MRI for the breast tumor using skin-affixed markers was performed immediately before excision. A percutaneous intramammary dye marker was applied to delineate an excision line, and the computer software 3D Slicer generated a real-time 3D virtual reality model of the tumor and the puncture needle in the breast. Excision of the tumor was performed in the usual manner along the excision line indicated with the dye. The resected specimens were carefully examined histopathologically. The mean mismatch between the navigation system and real distance was 2.01±0.32 mm when evaluated with the 3D phantom. Under guidance by the navigation system, a percutaneous intramammary dye marker was applied without any difficulty. Fiducial registration errors were 3.00 mm for patient no. 1, and 4.07 mm for patient no. 2. Histopathological examinations of the resected specimens of the 2 patients showed noninvasive ductal carcinoma in situ. The surgical margins were free of carcinoma cells. Real-time 3D virtual reality navigation system with open MRI is feasible for safe and accurate excision of nonpalpable MRI-detected breast tumors. Long-term outcomes of this technique should be evaluated further. |
 Left, percutaneous intramammary injection of dye in the syringe to make a dissection line of the tumor with the navigation of 3D Slicer. Arrowheads, skin markers; asterisk, a syringe attached with infrared markers; right, virtual MR images on the monitor of 3D Slicer reflecting the same slice as the puncture line. A breast tumor is indicated in green; a needle and a puncture line are indicated in red. |
Interfaces and Integration of Medical Image Analysis Frameworks: Challenges and Opportunities
|
Publication: Annu ORNL Biomed Sci Eng Cent Conf. 2010 May 25;2010:1-4. PMID: 21151892 | PDF Authors: Covington K, McCreedy ES, Chen M, Carass A, Aucoin N, Landman BA Institution: Department of Electrical Engineering, Vanderbilt University, Nashville, TN 37215 USA. Background/Purpose: Clinical research with medical imaging typically involves large-scale data analysis with interdependent software toolsets tied together in a processing workflow. Numerous, complementary platforms are available, but these are not readily compatible in terms of workflows or data formats. Both image scientists and clinical investigators could benefit from using the framework which is a most natural fit to the specific problem at hand, but pragmatic choices often dictate that a compromise platform is used for collaboration. Manual merging of platforms through carefully tuned scripts has been effective, but exceptionally time consuming and is not feasible for large-scale integration efforts. Hence, the benefits of innovation are constrained by platform dependence. Removing this constraint via integration of algorithms from one framework into another is the focus of this work. We propose and demonstrate a light-weight interface system to expose parameters across platforms and provide seamless integration. In this initial effort, we focus on four platforms Medical Image Analysis and Visualization (MIPAV), Java Image Science Toolkit (JIST), command line tools, and 3D Slicer. We explore three case studies: (1) providing a system for MIPAV to expose internal algorithms and utilize these algorithms within JIST, (2) exposing JIST modules through self-documenting command line interface for inclusion in scripting environments, and (3) detecting and using JIST modules in 3D Slicer. We review the challenges and opportunities for light-weight software integration both within development language (e.g., Java in MIPAV and JIST) and across languages (e.g., C/C++ in 3D Slicer and shell in command line tools). Funding:
|
Evaluation of Robotic Needle Steering in ex vivo Tissue
|
Publication: IEEE Int Conf Robot Autom. 2010 May 3;2010:2068-2073. PMID: 21339851 | PDF Authors: Majewicz A, Wedlick TR, Reed KB, Okamura AM. Institution: Department of Mechanical Engineering, Laboratory for Computational Sensing and Robotics, Johns Hopkins University, Baltimore, MD, USA. Background/Purpose: Insertion velocity, tip asymmetry, and shaft diameter may influence steerable needle insertion paths in soft tissue. In this paper we examine the effects of these variables on needle paths in ex vivo goat liver, and demonstrate practical applications of robotic needle steering for ablation, biopsy, and brachytherapy. All experiments were performed using a new portable needle steering robot that steers asymmetric-tip needles under fluoroscopic imaging. For bevel-tip needles, we found that larger diameter needles resulted in less curvature, i.e. less steerability, confirming previous experiments in artificial tissue. The needles steered with radii of curvature ranging from 3:4 cm (for the most steerable pre-bent needle) to 2:97m (for the least steerable bevel needle). Pre-bend angle significantly affected needle curvature, but bevel angle did not. We hypothesize that biological tissue characteristics such as inhomogeneity and viscoelasticity significantly increase path variability. These results underscore the need for closed-loop image guidance for needle steering in biological tissues with complex internal structure. Funding:
|
An open source implementation of colon CAD in 3D Slicer
|
Publication: Proc. of SPIE Vol. 7624 762421-2 | PDF Authors: Haiyong Xu, H. Donald Gageb, Pete Santago Institution: Department of Biomedical Engineering, Wake Forest Univ. Health Sciences, Medical Center Blvd. Winston-Salem, NC, USA. Background/Purpose: Most colon CAD (computer aided detection) software products, especially commercial products, are designed for use by radiologists in a clinical environment. Therefore, those features that effectively assist radiologists in finding polyps are emphasized in those tools. However, colon CAD researchers, many of whom are engineers or computer scientists, are working with CT studies in which polyps have already been identified using CT Colonography (CTC) and/or optical colonoscopy (OC). Their goal is to utilize that data to design a computer system that will identify all true polyps with no false positive detections. Therefore, they are more concerned with how to reduce false positives and to understand the behavior of the system than how to find polyps. Thus, colon CAD researchers have different requirements for tools not found in current CAD software. We have implemented a module in 3D Slicer to assist these researchers. As with clinical colon CAD implementations, the ability to promptly locate a polyp candidate in a 2D slice image and on a 3D colon surface is essential for researchers. Our software provides this capability, and uniquely, for each polyp candidate, the prediction value from a classifier is shown next to the 3D view of the polyp candidate, as well as its CTC/OC finding. This capability makes it easier to study each false positive detection and identify its causes. We describe features in our colon CAD system that meets researchers’ specific requirements. Our system uses an open source implementation of a 3D Slicer module, and the software is available to the public for use and for extension (http://www2.wfubmc.edu/ctc/download/).
|
In vivo visualization of cranial nerve pathways in humans using diffusion-based tractography
|
Publication: Neurosurgery. 2010 Apr;66(4):788-95; discussion 795-6. PMID: 20305498 | PDF Authors: M. Hodaie, J. Quan, D.Q.Chen Institution: Division of Neurosurgery, University of Toronto and Toronto Western Hospital, Toronto, Canada. Background/Purpose: Diffusion-based tractography has emerged as a powerful technique for 3-dimensional tract reconstruction and imaging of white matter fibers; however, tractography of the cranial nerves has not been well studied. In particular, the feasibility of tractography of the individual cranial nerves has not been previously assessed. METHODS: 3-Tesla magnetic resonance imaging scans, including anatomic magnetic resonance images and diffusion tensor images, were used for this study. Tractography of the cranial nerves was performed using 3D Slicer software. The reconstructed 3-dimensional tracts were overlaid onto anatomic images for determination of location and course of intracranial fibers. Detailed tractography of the cranial nerves was obtained, although not all cranial nerves were imaged with similar anatomic fidelity. Some tracts were imaged in great detail (cranial nerves II, III, and V). Tractography of the optic apparatus allowed tracing from the optic nerve to the occipital lobe, including Meyer's loop. Trigeminal tractography allowed visualization of the gasserian ganglion as well as postganglionic fibers. Tractography of cranial nerve III shows the course of the fibers through the midbrain. Lower cranial nerves (cranial nerves IX, XI, and XII) could not be imaged well. Tractography of the cranial nerves is feasible, although technical improvements are necessary to improve the tract reconstruction of the lower cranial nerves. Detailed assessment of anatomy and the ability of overlaying the tracts onto anatomic magnetic resonance imaging scans is essential, particularly in the posterior fossa, to ensure that the tracts have been reconstructed with anatomic fidelity. |
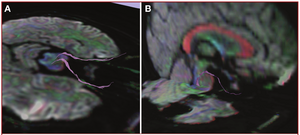 Intra- and extracranial course of cranial nerve III, depicted onto mixed anatomic/color-by-orientation MRI slices. Images are overlaid onto axial MRI slices (A) and sagittal MRI slices (B). The intracranial course takes a sharp angle, after which the fibers appear to enter the medial longitudinal fasciculus. The large commissural fibers of the corpus callosum (red) and fornix (blue) can be easily visualized on the sagittal slices. (Please see also Supplementary Digital Content 2, Figure 2, a 3-dimensional representation of the oculomotor nerves.) |
Coronary Artery Centerline Extraction in 3D Slicer using VMTK based Tools
|
Publication: Master's Thesis, University of Heidelberg 2010 Feb; | PDF Author: Daniel Hähn Institution: Medical Informatics, University of Heidelberg, Germany Background/Purpose: Atherosclerosis of the coronary arteries leads to Coronary Artery Disease (CAD) and is one of the main causes of death worldwide. Diagnosis and investigation of the severity of atherosclerosis is performed through medical imaging. In this context, the central lumen line of coronary arteries is of importance because it can be used for visualization and reconstruction based on reformatted images. We studied algorithms performing vessel enhancement filtering, level set segmentation and centerline computation based on Delaunay tessellation. By composing a pipeline of these, it is possible to extract the centerlines of coronary arteries in Computed Tomography Angiography (CTA) images. We implemented the pipeline as modules in 3D Slicer, an application providing a wide range of tools for medical image processing. This was accomplished by using methods of the Vascular Modeling Toolkit, an open source framework for image-based modeling of blood vessels. We evaluated the pipeline on eight CTA datasets by extracting the centerlines of the right and left coronary arteries. Using a large-scale parameter exploration study, we could achieve an average overlap over 72% of the clinically relevant part of the vessels combined with an average accuracy of 0.47mm by finding individual parameters for each dataset. Through a leave-one-out cross validation approach, we recognized parameter sets targeting the different branches of the arteries suitable for all datasets. Nevertheless, the found pattern lowers the average overlap to 54% and the average accuracy to 0.66mm. The software modules created during this project are available as open source software and suitable for end-users. Funding:
|
 A close-up of the internal Voronoi diagram and the corresponding centerline of a blood vessel obtained using the created tools in 3D Slicer. The individual Voronoi regions are visible as ”tiles” where the color indicates the maximum inscribed sphere radius R_M ( x ) [vx]. Surface noise leads to the small perturbations visible as spikes near the vessel lumen wall. |
Assessment of Image Registration Accuracy in Three-dimensional Transrectal Ultrasound Guided Prostate Biopsy
|
Publication: Med Phys. 2010 Feb;37(2):802-13. PMID: 20229890 Authors: Karnik V.V., Fenster A., Bax J., Cool D.W., Gardi L., Gyacskov I., Romagnoli C., Ward A.D. Institution: Biomedical Engineering Graduate Program, The University of Western Ontario, London, Ontario N6A 5C1, Canada. Background/Purpose: Prostate biopsy, performed using two-dimensional (2D) transrectal ultrasound (TRUS) guidance, is the clinical standard for a definitive diagnosis of prostate cancer. Histological analysis of the biopsies can reveal cancerous, noncancerous, or suspicious, possibly precancerous, tissue. During subsequent biopsy sessions, noncancerous regions should be avoided, and suspicious regions should be precisely rebiopsied, requiring accurate needle guidance. It is challenging to precisely guide a needle using 2D TRUS due to the limited anatomic information provided, and a three-dimensional (3D) record of biopsy locations for use in subsequent biopsy procedures cannot be collected. Our tracked, 3D TRUS-guided prostate biopsy system provides additional anatomic context and permits a 3D record of biopsies. However, targets determined based on a previous biopsy procedure must be transformed during the procedure to compensate for intraprocedure prostate shifting due to patient motion and prostate deformation due to transducer probe pressure. Thus, registration is a critically important step required to determine these transformations so that correspondence is maintained between the prebiopsied image and the real-time image. Registration must not only be performed accurately, but also quickly, since correction for prostate motion and deformation must be carried out during the biopsy procedure. The authors evaluated the accuracy, variability, and speed of several surface-based and image-based intrasession 3D-to-3D TRUS image registration techniques, for both rigid and nonrigid cases, to find the required transformations. Our surface-based rigid and nonrigid registrations of the prostate were performed using the iterative-closest-point algorithm and a thin-plate spline algorithm, respectively. For image-based rigid registration, the authors used a block matching approach, and for nonrigid registration, the authors define the moving image deformation using a regular, 3D grid of B-spline control points. The authors measured the target registration error (TRE) as the postregistration misalignment of 60 manually marked, corresponding intrinsic fiducials. The authors also measured the fiducial localization error (FLE), the effect of segmentation variability, and the effect of fiducial distance from the transducer probe tip. Lastly, the authors performed 3D principal component analysis (PCA) on the x, y, and z components of the TREs to examine the 95% confidence ellipsoids describing the errors for each registration method. Using surface-based registration, the authors found mean TREs of 2.13±0.80 and 2.09±0.77 mm for rigid and nonrigid techniques, respectively. Using image-based rigid and non-rigid registration, the authors found mean TREs of 1.74±0.84 and 1.50±0.83 mm, respectively. Our FLE was 0.21 mm and did not dominate the overall TRE. However, segmentation variability contributed substantially approximately 50%) to the TRE of the surface-based techniques. PCA showed that the 95% confidence ellipsoid encompassing fiducial distances between the source and target registration images was reduced from 3.05 to 0.14 cm3, and 0.05 cm3 for the surface-based and image-based techniques, respectively. The run times for both registration methods were comparable at less than 60 s. Our results compare favorably with a clinical need for a TRE of less than 2.5 mm, and suggest that image-based registration is superior to surface-based registration for 3D TRUS-guided prostate biopsies, since it does not require segmentation. |
Language laterality in autism spectrum disorder and typical controls: a functional, volumetric, and diffusion tensor MRI study
|
Publication: Brain Lang. 2010 Feb;112(2):113-20. PMID: 20031197 | PDF Authors: Knaus TA, Silver AM, Kennedy M, Lindgren KA, Dominick KC, Siegel J, Tager-Flusberg H. Institution: Department of Anatomy and Neurobiology, Boston University School of Medicine, Boston, MA, USA. Background/Purpose: Language and communication deficits are among the core features of autism spectrum disorder (ASD). Reduced or reversed asymmetry of language has been found in a number of disorders, including ASD. Studies of healthy adults have found an association between language laterality and anatomical measures but this has not been systematically investigated in ASD. The goal of this study was to examine differences in gray matter volume of perisylvian language regions, connections between language regions, and language abilities in individuals with typical left lateralized language compared to those with atypical (bilateral or right) asymmetry of language functions. Fourteen adolescent boys with ASD and 20 typically developing adolescent boys participated, including equal numbers of left- and right-handed individuals in each group. Participants with typical left lateralized language activation had smaller frontal language region volume and higher fractional anisotropy of the arcuate fasciculus compared to the group with atypical language laterality, across both ASD and control participants. The group with typical language asymmetry included the most right-handed controls and fewest left-handers with ASD. Atypical language laterality was more prevalent in the ASD than control group. These findings support an association between laterality of language function and language region anatomy. They also suggest anatomical differences may be more associated with variation in language laterality than specifically with ASD. Language laterality therefore may provide a novel way of subdividing samples, resulting in more homogenous groups for research into genetic and neurocognitive foundations of developmental disorders. |
Perk Station-Percutaneous Surgery Training and Performance Measurement Platform
|
Publication: Comput Med Imaging Graph. 2010 Jan;34(1):19-32. PMID: 19539446 | PDF Authors: Vikal S, U-Thainual P, Carrino JA, Iordachita I, Fischer GS, Fichtinger G. Institution: Queen's University, Kingston, ON, Canada. Background/Purpose: Image-guided percutaneous (through the skin) needle-based surgery has become part of routine clinical practice in performing procedures such as biopsies, injections and therapeutic implants. A novice physician typically performs needle interventions under the supervision of a senior physician; a slow and inherently subjective training process that lacks objective, quantitative assessment of the surgical skill and performance. Shortening the learning curve and increasing procedural consistency are important factors in assuring high-quality medical care. This paper describes a laboratory validation system, called Perk Station, for standardized training and performance measurement under different assistance techniques for needle-based surgical guidance systems. The initial goal of the Perk Station is to assess and compare different techniques: 2D image overlay, biplane laser guide, laser protractor and conventional freehand. The main focus of this manuscript is the planning and guidance software system developed on the 3D Slicer platform, a free, open source software package designed for visualization and analysis of medical image data. The prototype Perk Station has been successfully developed, the associated needle insertion phantoms were built, and the graphical user interface was fully implemented. The system was inaugurated in undergraduate teaching and a wide array of outreach activities. Initial results, experiences, ongoing activities and future plans are reported. Funding:
|
2009
Three-dimensional appearance of the lips muscles with three-dimensional isotropic MRI: in vivo study
|
Publication: Int J Comput Assist Radiol Surg. 2009 Jun;4(4):349-52. PMID: 20033581 | PDF Authors: R. Olszewski, Y. Liu, T. Duprez, T.M. Xu, H. Reychler Institution: Department of Oral and Maxillofacial Surgery, Cliniques Universitaires Saint Luc, Université Catholique de Louvain, Brussels, Belgium. Background/Purpose: Our knowledge of facial muscles is based primarily on atlases and cadaveric studies. This study describes a non-invasive in vivo method (3D MRI) for segmenting and reconstructing facial muscles in a three-dimensional fashion. Three-dimensional (3D), T1-weighted, 3 Tesla, isotropic MRI was applied to a subject. One observer performed semi-automatic segmentation using the Editor module from the 3D Slicer software (Harvard Medical School, Boston, MA, USA), version 3.2. We were able to successfully outline and three-dimensionally reconstruct the following facial muscles: pars labialis orbicularis oris, m. levatro labii superioris alaeque nasi, m. levator labii superioris, m. zygomaticus major and minor, m. depressor anguli oris, m. depressor labii inferioris, m. mentalis, m. buccinator, and m. orbicularis oculi. 3D reconstruction of the lip muscles should be taken into consideration in order to improve the accuracy and individualization of existing 3D facial soft tissue models. More studies are needed to further develop efficient methods for segmentation in this field. |
Traces
|
Author: Caitlin Berrigan Assistance: Marianna Jakab, BWH Special Thanks: J. Levi Schmidt (MIT), Tom Lutz (MIT Media Lab), Neil Gershenfeld (MIT Center for Bits & Atoms), Clare Tempany (BWH) Institution: MIT Visual Arts Program, 2010 Jun; Background/Purpose: Traces is a renewable sculpture of the artist’s own disembodied kidney, cast in frozen spit. Every two hours a new frozen organ is put on display, only to melt and drip away. The artist - using 3D Slicer - carefully traced the topography of her internal organ from a 3D MRI, consisting of hundreds of sequential medical images, in order to materialize its form outside of her body. Traces is a poetic deterritorialization of medical biotechnologies, organs without bodies and fleshy displacements. It calls attention to the alienability of body parts and the vast global industry that sustains the promise of an infinitely repairable body. Funding:
|
CLIMB study (Comprehensive Longitudinal Investigation of Multiple Sclerosis at Brigham and Women’s Hospital) former NHS study (The Harvard Multiple Sclerosis Natural History Study)
Steroid Adjunctive Treatment at Initiation of Avonex Therapy for Patients with Mono-Symptomatic or Relapsing-Remitting Multiple Sclerosis
|
Principal Investigator: Rohit Bakshi Institution: Laboratory for Neuroimaging Research, Partners MS Center Background/Purpose: We used quantitative MRI parameters such as T2 lesion volume (T2LV) and brain parenchymal fraction (BPF) to assess treatment in patients with MS. T2 lesion volume and BPF were measured using template-driven segmentation (TDS+) algorithm by automated pipeline. Medical image data were visualized by 3D Slicer to provide QC for original data, segmented output and perform manual correction on required steps of processing pipeline. Funding:
|
Age-Related Changes of Cognition in Health and Diseases: Image Analysis Core
|
Principal Investigator: Marilyn S. Albert Institution: Johns Hopkins Alzheimer’s Disease Research Center Background/Purpose: The goal of this project was the utilization of magnetic resonance imaging for morphometric analysis and tissue characterization to distinguish the patterns of brain atrophy and gray or white matter changes and white matter signal abnormalities in normal aging or Alzheimer's disease. Image analysis comprised an automated tissue segmentation into the intracranial cavity (ICC) and subclasses of white matter (WM), gray matter (GM), cerebrospinal fluid (CSF), and WM signal abnormalities (WMSA). Brain parenchymal fraction (BPF) was defined as BPF=(1-CSF)/ICC. Medical image data were visualized by 3D Slicer to provide QC for original grayscale data, segmented output and perform manual correction of ICC. Funding:
|
Harvard Research Nursing Home Project (MOBILIZE study)
|
Principal Investigator: Lewis A. Lipsitz Institution: Institute for Aging Research (IFAR) Background/Purpose: The project focuses on abnormal blood pressure regulation and its relationship to the development of falls and syncope in the elderly. 3D Slicer was used to provide visualization during QC step of original data (1.5T &3T), segmented output and perform manual correction on required steps of processing pipeline as well as corpus callosum outline for parcellation. |
Daclizumab Use in Patients with Pediatric Multiple Sclerosis Failing Interferon
|
Principal Investigator: Tanuja Chitnis Institution: Partners Pediatric MS Center Background/Purpose: We used quantitative MRI parameters such as T2 lesion volume (T2LV) to assess treatment in patients with POMS. The T2-lesion volume of the brain was manually outlined using 3D Slicer (www.slicer.org). Funding:
|
MRI findings of pediatric-onset Multiple Sclerosis patients: a retrospective study
|
Principal Investigator: Tanuja Chitnis Institution: Partners Pediatric MS Center Background/Purpose: To assess differences in MRI-derived lesion volume and brain parenchymal fraction (BPF) between patients with pediatric- and adult-onset Multiple Sclerosis (MS). T2 lesion volume and BPF were measured using template-driven segmentation (TDS+) algorithm by automated pipeline of the Multiple Sclerosis Database Project. Medical image data were visualized by 3D Slicer to provide QC for original data, segmented output and perform manual correction on required steps of processing pipeline. Funding:
|
Pediatric MS MRI Pilot Project
|
Principal Investigator: Tanuja Chitnis Institution: Partners Pediatric MS Center Background/Purpose: The goal of this project is to assess the characteristics of the pediatric MS patient population and increase education and awareness of this disease subtype. 3D Slicer is used to QC MRI images coming to the multi-center MRI data repository at the Center for Neurological Imaging (CNI) at Brigham and Women’s Hospital. Funding:
|
The Multiple Sclerosis Database Project (http://cni-boston.org)
|
Institution: Multiple Sclerosis Center Background/Purpose: Project is an initiative to design, develop and implement a database system integrating all aspects of clinical data with medical image data. This image-centric database will enable users to visualize clinical data, including laboratory results, immunologic data, and examination scores, simultaneously with that individual’s MRI images. By integrating the medical images and clinical patient data in a comprehensive system, the efficiency of patient care and data analysis is dramatically improved. This database is another small step towards our ultimate goal: Finding a cure for MS. Image Workflow system based on Standard Operation Procedures/Modules in Database system to allow the day-to-day management and query from data fields to ensure that trial was performed according to SOPs by managing the efforts of MRI research personnel, quality control of data, image analysis and image processing as a part of quantitative pipeline for trial. 3D Slicer (www.slicer.org) advanced image visualization package is used for visualization and analysis MRI data to provide T2 lesion volume and BPF volumetric measurements. |
Effects of Strain Thresholds on Bone Formation in Response to Mechanical Loading
|
Publication: TBA Authors: Ganesh Thiagarajan, Mark Johnson, Mark Dallas, Yunkai Lu Institution: University of Missouri-Kansas City Background/Purpose: It has been hypothesized that osteocytes are stimulated by local strain distribution within the bone subjected to mechanical loadings. In this research project, the mouse forearm model was built which was later used for extensive finite element study. The study will be the first in the field to include the radius in the numerical analysis to examine its effect on the load distribution between the ulna/radius. In order to preserve the geometrical details of the bones, the cavities were traced independently. Then a 3D volume subtraction was performed in another software to produce the exact CAD model of the bone structure. The results of the current research are expected to shed light on how bone perceives mechanical load and the pathway whereby the physical load is transduced into a biochemical event and eventually results in new bone formation. The study will help in developing new treatments for bone diseases such as osteoporosis. Funding:
|
Automated ventricular systems segmentation in brain CT images by combining low-level segmentation and high-level template matching
|
Publication: BMC Med Inform Decis Mak. 2009 Nov 3;9 Suppl 1:S4. PMID: 19891798 | PDF Authors: Chen W, Smith R, Ji SY, Ward KR, Najarian K. Institution: Department of Computer Science, Virginia Commonwealth University, Richmond, VA, USA. Background/Purpose: Accurate analysis of CT brain scans is vital for diagnosis and treatment of Traumatic Brain Injuries (TBI). Automatic processing of these CT brain scans could speed up the decision making process, lower the cost of healthcare, and reduce the chance of human error. In this paper, we focus on automatic processing of CT brain images to segment and identify the ventricular systems. The segmentation of ventricles provides quantitative measures on the changes of ventricles in the brain that form vital diagnosis information.
|
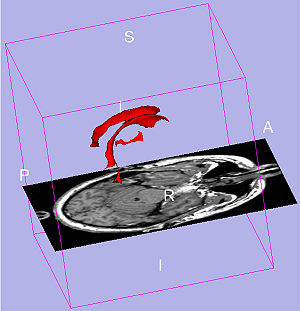 3D ventricle model from 3D Slicer sample visualization. The red parts are ventricular system model. Two symmetrical lateral ventricles on each side in normal case. The lower part in the middle is the third ventricle. The fourth ventricle is the lowest part which is not considered in our paper, but it can be included following the method in this paper. |
MRI-derived measurements of human subcortical, ventricular and intracranial brain volumes: Reliability effects of scan sessions, acquisition sequences, data analyses, scanner upgrade, scanner vendors and field strengths
|
Publication: Neuroimage. 2009 May 15;46(1):177-92. PMID: 19233293 | PDF Authors: Jovicich J, Czanner S, Han X, Salat D, van der Kouwe A, Quinn B, Pacheco J, Albert M, Killiany R, Blacker D, Maguire P, Rosas D, Makris N, Gollub R, Dale A, Dickerson BC, Fischl B. Institution: Center for Mind-Brain Sciences, Department of Cognitive and Education Sciences, University of Trento, Italy. Background/Purpose: Automated MRI-derived measurements of in-vivo human brain volumes provide novel insights into normal and abnormal neuroanatomy, but little is known about measurement reliability. Here we assess the impact of image acquisition variables (scan session, MRI sequence, scanner upgrade, vendor and field strengths), FreeSurfer segmentation pre-processing variables (image averaging, B1 field inhomogeneity correction) and segmentation analysis variables (probabilistic atlas) on resultant image segmentation volumes from older (n=15, mean age 69.5) and younger (both n=5, mean ages 34 and 36.5) healthy subjects. The variability between hippocampal, thalamic, caudate, putamen, lateral ventricular and total intracranial volume measures across sessions on the same scanner on different days is less than 4.3% for the older group and less than 2.3% for the younger group. Within-scanner measurements are remarkably reliable across scan sessions, being minimally affected by averaging of multiple acquisitions, B1 correction, acquisition sequence (MPRAGE vs. multi-echo-FLASH), major scanner upgrades (Sonata-Avanto, Trio-TrioTIM), and segmentation atlas (MPRAGE or multi-echo-FLASH). Volume measurements across platforms (Siemens Sonata vs. GE Signa) and field strengths (1.5 T vs. 3 T) result in a volume difference bias but with a comparable variance as that measured within-scanner, implying that multi-site studies may not necessarily require a much larger sample to detect a specific effect. These results suggest that volumes derived from automated segmentation of T1-weighted structural images are reliable measures within the same scanner platform, even after upgrades; however, combining data across platform and across field-strength introduces a bias that should be considered in the design of multi-site studies, such as clinical drug trials. The results derived from the young groups (scanner upgrade effects and B1 inhomogeneity correction effects) should be considered as preliminary and in need for further validation with a larger dataset. Fundings:
|
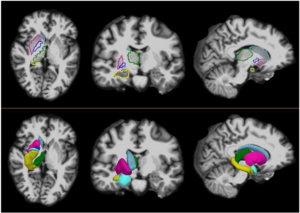 Sample color-coded subcortical segmentation results (right hemisphere only): hippocampus (yellow), thalamus (green), caudate (light blue), putamen (pink), pallidum (dark blue) and amygdala (turquoise). Top: Freesurfer derived subcortical labels, from a two-averaged MPRAGE in axial (left), coronal (center) and sagittal (right) views. Bottom: 3D surface models created with 3D Slicer derived from the Freesurfer subcortical segmentations. |
Integration of the Vascular Modeling Toolkit in 3D Slicer
|
Publication: Student Research Project 2009 Apr; | PDF Author: Daniel Hähn Institution: Medical Informatics, University of Heidelberg Background/Purpose: The extraction of vessels in two- and three-dimensional images is part of many clinical analysis tasks. Surgical and radiology procedures often involve the visualization and quantification of vessels in order to perform surgical planning or diagnostics. There is no single segmentation method that can extract vessels from every medical image modality, but different approaches and robust algorithms exist. Various published key algorithms are available within an open-source framework for image-based modeling of blood vessels, referred to as the Vascular Modeling Toolkit (VMTK). The library of VMTK was made available in 3D Slicer, an application providing a wide range of tools for medical image processing. This was realized using a hidden loadable module approach in order to provide a flexible way of distributing and including the library. To evaluate and verify the integration, a software module offering VMTK level set segmentation methods within 3D Slicer was created. With the successful connection of the two above mentioned software solutions, processing pipelines between VMTK code and other algorithms can be established. Several techniques for three dimensional reconstruction, geometric analysis, mesh generation and surface data analysis for image-based modeling of blood vessels are now accessible to the 3D Slicer developer. The reference implementation for accessing VMTK, as well as the created library module are available as open-source software. Funding:
|
New EMSegment Module in Slicer3
|
Publication: Insight Journal, Sep 15, 2009. Authors: Nicolas Rannou1, Sylvain Jaume2, Steve Pieper3, Ron Kikinis3 Institution: 1ISEN Brest (France), 2MIT CSAIL, 3Harvard Medical School BWH Background/Purpose: Many neuroanatomy studies rely on brain tissue segmentation in Magnetic Resonance images (MRI). The Expectation-Maximization (EM) theory offers a popular framework for this task. We studied the EM algorithm developed at the Surgical Planning Laboratory (SPL) at Harvard Medical School and implemented in the Slicer3 software. We observed that the segmentation lacks accuracy if the image exhibits some intensity inhomogeneity. Moreover the optimum parameters are challenging to estimate. This document aims at describing our solutions within the context of statistical modeling. Our contributions range from algorithm improvements to novel representations of the statistical distribution model. First we added a bias field correction module and exposed the most significant parameters. Second we proposed a new way to select the distribution of the tissues to be segmented. Finally we designed a set of interactive tools to make the segmentation process easier and more accurate. To validate the new segmentation pipeline, we performed our experiments on MRI data and a clinical expert evaluated our results. Funding:
|
 New EMSegment Module in Slicer3. We improved the usability of EMSegment in a clinical setting and studied the influence of MRI Bias Field Correction on the segmentation accuracy. |
|
Publication: The International Journal of Virtual Reality, 2009, 8(1): 9-16. | PDF Authors: Huy Hoang Tran, Kiyoshi Matsumiya, Ken Masamune, Ichiro Sakuma, Takeyoshi Dohi and Hongen Liao Institution: Graduate School of Information Science and Technology, The University of Tokyo, Tokyo, Japan. Background/Purpose: In this paper, we mainly focus on a high-speed IV rendering method that allows interactions to be done in almost real-time, while maintaining the rendered result at an acceptable quality. We also built a navigation system based on the open source software 3D Slicer so that many tasks can be done in a single application. Funding:
|
Smaller Amygdala is Associated with Anxiety in Patients with Panic Disorder
|
Publication: Psychiatry and Clinical Neurosciences 2009; 63: 266-276. PMID: 19566756 | PDF Authors: Hayano F, Nakamura M, Asami T, Uehara K, Yoshida T, Roppongi T, Otsuka T, Inoue T, Hirayasu Y Institution: Department of Psychiatry, Yokohama City University, Yokohama, Kanagawa, Japan. Background/Purpose: Anxiety a core feature of panic disorder, is linked to function of the amygdala. Volume alterations in the brain of patients with panic disorder have previously been reported, but there has been no report of amygdala volume association with anxiety. METHODS: Volumes of hippocampus and amygdala were manually measured using magnetic resonance imaging obtained from 27 patients with panic disorder and 30 healthy comparison subjects. In addition the amygdala was focused on, applying small volume correction to optimized voxel-based morphometry (VBM). State-Trait Anxiety Inventory and the NEO Personality Inventory Revised were also used to evaluate anxiety. Amygdala volumes in both hemispheres were significantly smaller in patients with panic disorder compared with control subjects (left: t = -2.248, d.f. = 55, P = 0.029; right: t = -2.892, d.f. = 55, P = 0.005). VBM showed that structural alteration in the panic disorder group occurred on the corticomedial nuclear group within the right amygdala (coordinates [x,y,z (mm)]: [26,-6,-16], Z score = 3.92, family-wise error-corrected P = 0.002). The state anxiety was negatively correlated with the left amygdala volume in patients with panic disorder (r = -0.545, P = 0.016). These findings suggested that the smaller volume of the amygdala may be associated with anxiety in panic disorder. Of note, the smaller subregion in the amygdala estimated on VBM could correspond to the corticomedial nuclear group including the central nucleus, which may play a crucial role in panic attack. Funding:
|
 Voxels with a significant decrease in region of amygdala area among panic disorder (PD) patients compared with healthy control (HC) subjects. (a) statistical parametric map showing significantly smaller subregion within the amygdala. (b) The same statistical parametric map overlayed on a structural volume. |
Optimal Transseptal Puncture Location for Robot-Assisted Left Atrial Catheter Ablation
|
Publication: Int Conf Med Image Comput Comput Assist Interv. 2009;12(Pt 1): 1–8. PMID: 20425964 | PDF. Authors: Jagadeesan Jayender, Rajni V. Patel, Gregory F. Michaud, Nobuhiko Hata Institution: Harvard Medical School, Brigham and Women’s Hospital, Boston, MA, USA and The University of Western Ontario, Canadian Surgical Technologies and Advanced Robotics (CSTAR), London, ON, Canada. Background/Purpose: The preferred method of treatment for Atrial Fibrillation (AF) is by catheter ablation wherein a catheter is guided into the left atrium through a transseptal puncture. However, the transseptal puncture constrains the catheter, thereby limiting its maneuverability and increasing the difficulty in reaching various locations in the left atrium. In this project, we have developed a computational algorithm to choose the optimal transseptal puncture location to obtain maximum maneuverability of the catheter for performing cardiac ablation. We have employed an optimization algorithm to maximize the Global Isotropy Index (GII) to evaluate the optimal transseptal puncture location. Preoperative MR/CT images of the heart are segmented using the open source image-guided therapy software, 3D Slicer, to obtain models of the left atrium and septal wall. These models are input to the optimization algorithm to evaluate the optimal transseptal puncture location. The results of the GII algorithm are color-mapped on the model of the septal wall and displayed to the user in 3D Slicer. Funding:
|
 The result of the GII algorithm displayed as a color map on the septal wall. The color blue represents the GII with highest value, which correspond to the optimal port, while red represents a low value of GII. Discrete points on the septal wall which provide the maximum maneuverability of the catheter within the left atrium. |
Measurements from image-based three dimensional pelvic floor reconstruction: a study of inter- and intraobserver reliability
|
Publication: J Magn Reson Imaging. 2009 Aug;30(2):344-50. PMID: 19629987 | PDF Authors: Hoyte L, Brubaker L, Fielding JR, Lockhart ME, Heilbrun ME, Salomon CG, Ye W, Brown MB; Pelvic Floor Disorders Network. Institution: University of South Florida, College of Medicine, Division of Urogynecology and Pelvic Reconstructive Surgery, Tampa General Hospital, Urogynecology Division, Tampa, FL, USA. Background/Purpose: To describe inter- and intraobserver reliability of 3D measurements of female pelvic floor structures. Twenty reconstructed MR datasets of primiparas at 6-12 months postpartum were analyzed. Pelvic organ measurements were independently made twice by three radiologists blinded to dataset order. A "within-reader" analysis, a "between-reader" analysis, and the intraclass correlation (ICC), and standard deviation ratio (SDR) were computed for each parameter. Fifteen continuous variables and one categorical variable were measured. Eight continuous parameters showed excellent agreement (ICC >0.85 / SDR <0.40), five parameters showed relatively good agreement (ICC >0.70 / SDR >or=0.40, <0.60). Two parameters showed poor agreement (ICC <or=0.70 and/or SDR >or=0.60). The categorical variable showed poor agreement. Agreement was best where landmark edges were well defined, acceptable where more "reader judgment" was needed, and poor where levator defects made landmarks difficult to identify. Automated measurement algorithms are under study and may improve agreement in the future. Funding:
|
Three-dimensional analysis of rodent paranasal sinus cavities from X-ray computed tomography (CT) scans
|
Publication: Can J Vet Res. 2009 Jul;73(3):205-11. PMID: 19794893 | PDF. Authors: Jonathan E. Phillips, Lunan Ji, Maria A. Rivelli, Richard W. Chapman, Michel R. Corboz. Institution: Pulmonary and Peripheral Neurobiology, Schering-Plough Research Institute, 2015 Galloping Hill Road, Kenilworth, New Jersey 07033, USA. Background/Purpose: Continuous isometric microfocal X-ray computed tomography (CT) scans were acquired from an AKR/J mouse, Brown-Norway rat, and Hartley guinea pig. The anatomy and volume of the paranasal sinus cavities were defined from 2-dimensional (2-D) and 3-dimensional (3-D) CT images. Realistic 3-D images were reconstructed and used to determine the anterior maxillary, posterior maxillary, and ethmoid sinus cavity airspace volumes (mouse: 0.6, 0.7, and 0.7 mm 3, rat: 8.6, 7.7, and 7.0 mm3, guinea pig: 63.5, 46.6 mm3, and no ethmoid cavity, respectively). The mouse paranasal sinus cavities are similar to the corresponding rat cavities, with a reduction in size, while the corresponding maxillary sinus cavities in the guinea pig are different in size, location, and architecture. Also, the ethmoid sinus cavity is connected by a common drainage pathway to the posterior maxillary sinus in mouse and rat while a similar ethmoid sinus was not present in the guinea pig. We conclude that paranasal sinus cavity airspace opacity (2-D) or volume (3-D) determined by micro-CT scanning may be used to conduct longitudinal studies on the patency of the maxillary sinus cavities of rodents. This represents a potentially useful endpoint for developing and testing drugs in a small animal model of sinusitis. Funding:
|
 Volumetric surface renderings of paranasal sinus created from sequential computed tomography (CT) images. Three different orientations of the paranasal sinuses (anterior maxillary in green, posterior maxillary in blue, and anterior ethmoid in red) are shown with respect to location in skull according to species, all at different magnifications. |
Multimodal imaging in mild cognitive impairment: Metabolism, morphometry and diffusion of the temporal–parietal memory network
|
Publication: Neuroimage. 2009 Mar 1;45(1):215-23. PMID: 19056499 | PDF Authors: K.B. Walhovd, A.M. Fjell, I. Amlien, R. Grambaite, V. Stenset, A. Bjørnerud, I. Reinvang, L. Gjerstad, T. Cappelen, P. Due-Tønnessen, T. Fladby Institution: Center for the Study of Human Cognition, Department of Psychology, University of Oslo, Norway. Background/Purpose: This study compared sensitivity of FDG-PET, MR morphometry, and diffusion tensor imaging (DTI) derived fractional anisotropy (FA) measures to diagnosis and memory function in mild cognitive impairment (MCI). Patients (n = 44) and normal controls (NC, n = 22) underwent FDG-PET and MRI scanning yielding measures of metabolism, morphometry and FA in nine temporal and parietal areas affected by Alzheimer's disease and involved in the episodic memory network. Patients also underwent memory testing (RAVLT). Logistic regression analysis yielded 100% diagnostic accuracy when all methods and ROIs were combined, but none of the variables then served as unique predictors. Within separate ROIs, diagnostic accuracy for the methods combined ranged from 65.6% (parahippocampal gyrus) to 73.4 (inferior parietal cortex). Morphometry predicted diagnostic group for most ROIs. PET and FA did not uniquely predict group, but a trend was seen for the precuneus metabolism. For the MCI group, stepwise regression analyses predicting memory scores were performed with the same methods and ROIs. Hippocampal volume and FA of the retrosplenial WM predicted learning, and hippocampal metabolism and parahippocampal cortical thickness predicted 5 minute recall. No variable predicted 30 minute recall independently of learning. In conclusion, higher diagnostic accuracy was achieved when multiple methods and ROIs were combined, but morphometry showed superior diagnostic sensitivity. Metabolism, morphometry and FA all uniquely explained memory performance, making a multi-modal approach superior. Memory variation in MCI is likely related to conversion risk, and the results indicate potential for improved predictive power by the use of multimodal imaging. Funding:
|
 Illustrations of the main steps in the multi-modal image analysis. (A and B) The brain surface is parcellated into 34 different regions in each hemisphere (B), and a newly developed algorithm assigns a label to each underlying WM voxel (A). (C) Five cortical ROI's and hippocampus, of much importance in the episodic memory network, were chosen for analyses in the present paper. EC — entorhinal cortex; PH — parahippocampal cortex; RC — retrosplenial cortex; PC — posterior cingulate; Pre — precuneus. (D) Every voxel in each brain volume is assigned a label based on the cortical parcellations (A), the WM parcellations (B), and the whole-brain segmentation. The distance between the red and the yellow line is the cerebral cortex. (E) The FA volume is registered to the anatomical volume, and mean FA is calculated from the voxels included in each WM ROI (A). (F) FDG-PET data are also registered to the anatomical volume, and the metabolism is divided by the metabolism in the brainstem. Mean metabolism in each cortical ROI and hippocampus is calculated. (G) The whole-brain segmentation yields hippocampal volume (hippocampus is marked in yellow, indicated by the red arrows). A three-dimensional rendering of hippocampus is shown to illustrate the result of the segmentation. |
The relation between connection length and degree of connectivity in young adults: a DTI analysis
|
Publication: Cereb Cortex. 2009 Mar;19(3):554-62. PMID: 18552356 | PDF Authors: Lewis JD, Theilmann RJ, Sereno MI, Townsend J. Institution: Department of Cognitive Science, University of California-San Diego, La Jolla, CA, USA. Background/Purpose: Using diffusion tensor imaging and tractography to detail the patterns of interhemispheric connectivity and to determine the length of the connections, and formulae based on histological results to estimate degree of connectivity, we show that connection length is negatively correlated with degree of connectivity in the normal adult brain. The degree of interhemispheric connectivity--the ratio of interhemispheric connections to total corticocortical projection neurons--was estimated for each of 5 subregions of the corpus callosum in 22 normal males between 20 and 45 years of age (mean 31.68; standard deviation 8.75), and the average length of the longest tracts passing through each point of each subregion was calculated. Regression analyses were used to assess the relation between connection length and the degree of connectivity. Connection length was negatively correlated with degree of connectivity in all 5 subregions, and the regression was significant in 4 of the 5, with an average r(2) of 0.255. This is contrasted with previous analyses of the relation between brain size and connectivity, and connection length is shown to be a superior predictor. The results support the hypothesis that cortical networks are optimized to reduce conduction delays and cellular costs. Funding:
|
The ROBOCAST project: ROBOt and sensors integration for Computer Assisted Surgery and Therapy
|
Authors: Reuben R Shamir, Leo Joskowicz, Giancarlo Ferrigno Institution: School of Engineering and Computer Science, The Hebrew University of Jerusalem, Israel. Background/Purpose: The goal of the European FP7 ROBOCAST project is to develop a system for keyhole neurosurgery. ROBOCAST integrates optical and electromagnetic localization devices with two modified commercial surgical robots: 1. PathFinder (Prosurgics Ltd, UK), which is used for gross positioning, and; 2. MARS (Mazor Surgical Technologies Ltd, Israel), which is used for fine positioning. A third snake-like robot, currently under development, will be attached to the MARS robot and is intended for curved paths. The system includes as well a haptic device, a tracked ultrasound probe and a micro Doppler ultrasound probe. This sensor and actuation redundancy is expected to yield higher accuracy and robustness, to significantly increase the patient safety, and to allow new procedures. The ROBOCAST software is based on Slicer 3.4 and uses many of its functions. Funding:
|
A trajectory planning method for reduced patient risk in image-guided neurosurgery: preliminary results
|
Authors: Reuben R. Shamir, Ran Roth, Leo Joskowicz, Luca Antiga, Roberto I. Foroni, Yigal Shoshan Institution: School of Engineering and Computer Science, The Hebrew University of Jerusalem, Israel. Background/Purpose: We present a new preoperative planning method to better assess and reduce the risk caused by a misplacement of surgical tools in image-guided keyhole neurosurgery. The goal is to quantify the risk of a proposed straight trajectory and to find the trajectory with the lowest risk to hit nearby brain structures based on pre-operative CT/MRI images and their segmentation. First, distance maps are computed on the segmented data such that each voxel on the preoperative CT/MRI image is associated with a vector of distances to each of the segmented structures. A risk volume is then created from the distance maps such that a risk ratio is computed based on the vector of distances associated with each voxel. Next, the risk for each possible trajectory is computed such that the risk along a trajectory is assigned with the maximal risk ratio of intersected voxels. The trajectory that is associated with the lowest risk is presented using Slicer 3.2 to the neurosurgeon for further revision. The neurosurgeon can revise the suggested trajectory and can add new paths using interactive 3D visualization and quantitative risk assessment. The interactive 3D visualization shows the segmented structures to be avoided along with a geometric structure that envelopes the defined trajectory and represents the localization uncertainty. In addition, candidate entry points are represented by the outer head surface that is extracted automatically from the patient’s preoperative image and the neurosurgeon can select parts of the surface to restrict the optional entry points. A table, called ‘risk card’, of quantitative measures regarding the risk that is associated with the defined trajectory is presented. It shows the neurosurgeon meaningful geometrical information, including the path length and distance of segmented structures to the path. For the initial validation, we have compared the conventional approach for trajectory planning with our method on 8 targets selected at various locations on a clinical Magnetic Resonance Angiography (MRA) head image. For each target, the user selected two trajectories: one with the conventional method based on the axial, sagittal, and coronal 2D views of the original MRA image, and the second with our method. Our preliminary results show a significant reduction in trajectory risk and planning time when compared to the conventional method. Funding:
|
Contrast-maximizing adaptive region growing for CT
|
Publication: Poster Presentation at the Second International Workshop on Pulmonary Image Analysis. | Poster, Paper. Oral Presentation at Advanced Concepts for Intelligent Vision Systems, ACIVS 2009. Authors: Carlos S. Mendoza, Begoña Acha, Carmen Serrano. Institution: University of Sevilla, Signal Processing Department. Background/Purpose: We have developed a general segmentation framework based on a contrast-based self-assessed region growing strategy. Thanks to a previous normalization stage, a wide variety of CT imaging conditions are supported, requiring limited user intervention. Although multiple seeds are accepted for performance boosting, only a single-seed selection stage is required for most of our application specific tissues. The detection of the optimal parameters is managed internally using a measure of the varying contrast of the growing region. Validation is provided for some synthetic images, and also for real CT images. Results have been obtained in the context of surgical planning (bone, muscle and fat segmentation) and pulmonary image analysis (airway segmentation). Funding:
|
Computer-aided 3D visualization in oto-rhino-laryngology
|
Publication: Poster Presentation at the 80th Conference of the German ENT Society. | PDF. Authors: A. L. Nagy, A. Tanács, A. Czesznak, Gy. Smehák, F. Tóth, L. Rovó, J. Jóri, J. G. Kiss Institution: University of Szeged, Department of Oto-rhino-laryngology and Head, Neck Surgery Background/Purpose: Most of the CT and MR data is stored nowadays on digital media. This opens up new possibilities for us to inspect these data. In our department we evaluated the use of the 3D Slicer software, which is free, open source and is being actively developed. In oto-rhino-laryngology we often encounter situations where the exact localization and estimation of the size of a lesion, or tumor is of critical importance. Especially in the neck and the facial region an extended resection may easily cause damage to the function of the affected organ. The preservation of the swallowing function in case of the neck, and the preservation of sight in interventions around the orbita are two of these areas. By visualizing these space-occupying lesions we were able to plan and carry out the surgery more precisely. |
Clinical Application of Curvilinear Distraction Osteogenesis for Correction of Mandibular Deformities
|
Publication: J Oral Maxillofac Surg. 2009 May;67(5):996-1008. PMID: 19375009 | PDF Authors: Kaban LB, Seldin EB, Kikinis R, Yeshwant K, Padwa BL, Troulis MJ. Institution: Department of Oral and Maxillofacial Surgery, Massachusetts General Hospital, Harvard School of Dental Medicine, Boston, MA, USA. Background/Purpose: To report the use of a semiburied curvilinear distraction device, with a 3-dimensional (3D) computed tomography treatment planning system, for correction of mandibular deformities. This was a retrospective evaluation of 13 consecutive patients, with syndromic and nonsyndromic micrognathia, who underwent correction by curvilinear distraction osteogenesis. A 3D computed tomography scan was obtained for each patient and imported into a 3D treatment planning system (Slicer/Osteoplan). Surgical guides were constructed to localize the osteotomy and to drill holes to secure the distractor's proximal and distal footplates to the mandible. Postoperatively, patients were followed by clinical examination and plain radiographs to ensure the desired vector of movement. At end distraction, when possible, a 3D computed tomography scan was obtained to document the final mandibular position. Of the 13 patients, 8 were females and 5 were males, with a mean age of 11.9 years (range 15 months to 39 years). All 13 underwent bilateral mandibular curvilinear distraction. Of the 13 patients, 8 were 16 years old or younger and 5 were younger than 6 years of age. The diagnoses included Treacher Collins syndrome (n = 3), Nager syndrome (n = 3), craniofacial microsomia (n = 2), post-traumatic ankylosis (n = 1), and micrognathia (syndromic, n = 3; nonsyndromic, n = 1). The correct distractor placement, vector of movement, and final mandibular position were achieved in 10 of 13 patients. In the other 3 patients, the desired jaw position was achieved by "molding" the regenerate. The use of a semiburied curvilinear distraction device, with 3D treatment planning, is a potentially powerful tool to correct complex mandibular deformities. Funding:
|
 A, Preoperative and B, end-distraction lateral cephalograms. (Patient in Fig 4.) Posterior airway space increased from 5.03 to 18.96 mm and distance from hyoid to mandible decreased from 19.3 to 12.9 mm (normal values 11 mm and Ͻ15 mm, respectively). Overlay of predicted (blue) and end-fixation (pink) position of C, right and D, left sides of mandible demonstrating desired movement was achieved. |
In Vivo Hippocampal Measurement and Memory: A Comparison of Manual Tracing and Automated Segmentation in a Large Community-Based Sample
|
Publication: PLoS One. 2009 Apr; 4(4):e5265. PMID: 19370155 | PDF Authors: Cherbuin N1, Anstey KJ1, Réglade-Meslin C1, Sachdev PS2,3. Institution: 1Centre for Mental Health Research, Australian National University, Canberra, Australian Capital Territory, Australia. 2School of Psychiatry, University of New South Wales, Sydney, New South Wales, Australia. 3Neuropsychiatric Institute, Prince of Wales Hospital, Sydney, New South Wales, Australia. Background/Purpose: Each segmented volume was inspected by producing a 3D model using the 3D Slicer software package (www.slicer.org) and its quality rated: a) no or minor defects (estimated to be smaller than ,0.5% of total volume) b) moderate defects (,0.5–5% of total volume), major defects (greater than 5% of total volume). Exemplars are given in Figure 1. Funding:
|
How volumetric analysis quantifies therapeutic response of slow-flow vascular malformations
|
Publication: ECR Today 2009 March 9/10:23. | PDF. Institution: Medical University of Vienna, Austria. Authors: T. Dobrocky, H. Kubiena, R. Kikinis, J. Kettenbach Background/Purpose: Slow-flow vascular malformations, such as venous and lymphatic lesions, are most often recognised in infancy and are thus characterised by a rapid expansion. The majority of these lesions are found in the head and neck region, where many options for treatment modalities (drug therapy, percutaneous sclerotherapy, surgical resection) have been reported in the literature. In many cases, excellent results have been achieved by ultrasound-guided or fluoroscopy-guided percutaneous sclerotherapy, sometimes combined with final surgical resection (see figure). Diagnostic MRI of vascular malformations allows non-invasive treatment planning and follow-up, with excellent soft-tissue contrast. |
A minimally invasive registration method using surface template-assisted marker positioning (STAMP) for image-guided otologic surgery
|
Publication: Otolaryngol Head Neck Surg. 2009 Jan;140(1):96-102. PMID: 19130970 | PDF Authors: Matsumoto N, Hong J, Hashizume M, Komune S. Institution: Department of Otorhinolaryngology, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan. Background/Purpose: A new, minimally invasive registration method was developed for image-guided otologic surgery. We utilized laser-sintered template of the patient's bone surface to transfer the virtual markers to the patient's bone intraoperatively and eliminated the necessity for preoperative marker positioning or additional CT scan. Simulation surgeries and clinical application. We measured registration errors in 10 trials using replicas and six ear surgeries (two cochlear implant insertions, four translabyrinthine acoustic tumor removals). The target registration errors varied among the surgical targets. Errors were less than 1 mm near the cochlear implant insertion target both in phantom study and in actual surgeries. Our newly developed method reduced the preoperative procedures for patients but did not reduce the accuracy in cochlear implant surgery. Our method would be a useful image-guided surgery method in the field of otology, where both accuracy and noninvasiveness are required. Funding:
|
 The IGS screen during surgery. (A) The navigation view of the surgery during cochleostomy. Red, facial nerve; light green, semicircular canals; orange, cochlea; green, drill. (B) The navigation view of the surgery during translabyrinthine approach to the internal auditory canal. The surgeon is now pointing to target “SA” of Table 2. Red, facial nerve; orange, semicircular canals; light green, internal auditory canal; pink, sigmoid sinus and jugular bulb; green, drill with 10-mm extension marks. |
Diffusion Tractography of the Fornix in Schizophrenia
|
Publication: Schizophr Res. 2009 Jan;107(1):39-46. PMID: 19046624 | PDF Authors: Fitzsimmons J, Kubicki M, Smith K, Bushell G, Estepar RS, Westin CF, Nestor PG, Niznikiewicz MA, Kikinis R, McCarley RW, Shenton ME. Institution: Psychiatry Neuroimaging Laboratory, Department of Psychiatry, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA. Background/Purpose: White matter fiber tracts, especially those interconnecting the frontal and temporal lobes, are likely implicated in pathophysiology of schizophrenia. Very few studies, however, have focused on the fornix, a compact bundle of white matter fibers, projecting from the hippocampus to the septum, anterior nucleus of the thalamus and the mamillary bodies. Diffusion Tensor Imaging (DTI), and a new post-processing method, fiber tractography, provides a unique opportunity to visualize and to quantify entire trajectories of fiber bundles, such as the fornix, in vivo. We applied these techniques to quantify fornix diffusion anisotropy in schizophrenia. DTI images were used to evaluate the left and the right fornix in 36 male patients diagnosed with chronic schizophrenia and 35 male healthy individuals, group matched on age, parental socioeconomic status, and handedness. Regions of interest were drawn manually, blind to group membership, to guide tractography, and fractional anisotropy (FA), a measure of fiber integrity, was calculated and averaged over the entire tract for each subject. The Doors and People test (DPT) was used to evaluate visual and verbal memory, combined recall and combined recognition. Analysis of variance was performed and findings demonstrated a difference between patients with schizophrenia and controls for fornix FA (p=0.006). Protected post-hoc independent sample t-tests demonstrated a bilateral FA decrease in schizophrenia, compared with control subjects (left side: p=0.048; right side p=0.006). Higher fornix FA was statistically significantly correlated with DPT and measures of combined visual memory (r=0.554, p=0.026), combined verbal memory (r=0.647, p=0.007), combined recall (r=0.516, p=0.041), and combined recognition (r=0.710, p=0.002) for the control group. No such statistically significant correlations were found in the patient group. Our findings show the utility of applying DTI and tractography to study white matter fiber tracts in vivo in schizophrenia. Specifically, we observed a bilateral disruption in fornix integrity in schizophrenia, thus broadening our understanding of the pathophysiology of this disease. Funding:
|
Spiny Versus Stubby: 3D Reconstruction of Human Myenteric (type I) Neurons
|
Publication: Histochem Cell Biol. 2009 Jan;131(1):1-12. PMID: 18807064 | PDF Authors: Lindig TM, Kumar V, Kikinis R, Pieper S, Schrödl F, Neuhuber WL, Brehmer A. Institution: Department of Anatomy I, University of Erlangen-Nuremberg, Erlangen, Germany. Background/Purpose: We have compared the three-dimensional (3D) morphology of stubby and spiny neurons derived from the human small intestine. After immunohistochemical triple staining for leu-enkephalin (ENK), vasoactive intestinal peptide (VIP) and neurofilament (NF), neurons were selected and scanned based on their immunoreactivity, whether ENK (stubby) or VIP (spiny). For the 3D reconstruction, we focused on confocal data pre-processing with intensity drop correction, non-blind deconvolution, an additional compression procedure in z-direction, and optimizing segmentation reliability. 3D Slicer software enabled a semi-automated segmentation based on an objective threshold (interrater and intrarater reliability, both 0.99). We found that most dendrites of stubby neurons emerged only from the somal circumference, whereas in spiny neurons, they also emerged from the luminal somal surface. In most neurons, the nucleus was positioned abluminally in its soma. The volumes of spiny neurons were significantly larger than those of stubby neurons (total mean of stubbies 806±128 μm3, of spinies 2,316±545 μm3), and spiny neurons had more dendrites (26.3 vs. 11.3). The ratios of somal versus dendritic volumes were 1:1.2 in spiny and 1:0.3 in stubby neurons. In conclusion, 3D reconstruction revealed new differences between stubby and spiny neurons and allowed estimations of volumetric data of these neuron populations. Funding:
|
2008
|
Publication: Int J Med Robot. 2008 Dec;4(4):321-30. PMID: 18803337 | PDF Authors: Xia T, Baird C, Jallo G, Hayes K, Nakajima N, Hata N, Kazanzides P. Institution: Department of Computer Science, Johns Hopkins University, Baltimore, MD, USA. Background/Purpose: We developed an image-guided robot system to provide mechanical assistance for skull base drilling, which is performed to gain access for some neurosurgical interventions, such as tumour resection. The motivation for introducing this robot was to improve safety by preventing the surgeon from accidentally damaging critical neurovascular structures during the drilling procedure. We integrated a Stealthstation((R)) navigation system, a NeuroMate((R)) robotic arm with a six-degree-of-freedom force sensor, and the 3D Slicer visualization software to allow the robotic arm to be used in a navigated, cooperatively-controlled fashion by the surgeon. We employed virtual fixtures to constrain the motion of the robot-held cutting tool, so that it remained in the safe zone that was defined on a preoperative CT scan. We performed experiments on both foam skull and cadaver heads. The results for foam blocks cut using different registrations yielded an average placement error of 0.6 mm and an average dimensional error of 0.6 mm. We drilled the posterior porus acusticus in three cadaver heads and concluded that the robot-assisted procedure is clinically feasible and provides some ergonomic benefits, such as stabilizing the drill. We obtained postoperative CT scans of the cadaver heads to assess the accuracy and found that some bone outside the virtual fixture boundary was cut. The typical overcut was 1-2 mm, with a maximum overcut of about 3 mm. The image-guided cooperatively-controlled robot system can improve the safety and ergonomics of skull base drilling by stabilizing the drill and enforcing virtual fixtures to protect critical neurovascular structures. The next step is to improve the accuracy so that the overcut can be reduced to a more clinically acceptable value of about 1 mm. Funding:
|
The relationship between diffusion tensor imaging and volumetry as measures of white matter properties
|
Publication: Neuroimage. 2008 Oct 1;42(4):1654-68. PMID: 18620064 | PDF Authors: Anders M. Fjell, Lars T. Westlye, Doug N. Greve, Bruce Fischl, Thomas Benner, André J.W. van der Kouwe, David Salat, Atle Bjørnerud, Paulina Due-Tønnessen, Kristine B. Walhovd Institution: Center for the Study of Human Cognition, Department of Psychology, University of Oslo, Norway. Background/Purpose:There is still limited knowledge about the relationship between different structural brain parameters, despite huge progress in analysis of neuroimaging data. The aim of the present study was to test the relationship between fractional anisotropy (FA) from diffusion tensor imaging (DTI) and regional white matter (WM) volume. As WM volume has been shown to develop until middle age, the focus was on changes in WM properties in the age range of 40 to 60 years. 100 participants were scanned with magnetic resonance imaging (MRI). Each hemisphere was parcellated into 35 WM regions, and volume, FA, axial, and radial diffusion in each region were calculated. The relationships between age and the regional measures of FA and WM volume were tested, and then FA and WM volume were correlated, corrected for intracranial volume, age, and sex. WM volume was weakly related to age, while FA correlated negatively with age in 26 of 70 regions, caused by a mix of reduced axial and increased radial diffusion with age. 23 relationships between FA and WM volume were found, with seven being positive and sixteen negative. The positive correlations were mainly caused by increased radial diffusion. It is concluded that FA is more sensitive than volume to changes in WM integrity during middle age, and that FA-age correlations probably are related to reduced amount of myelin with increasing age. Further, FA and WM volume are moderately to weakly related and to a large extent sensitive to different characteristics of WM integrity. Funding:
|
 3D renderings of the probabilistic tracts overlaid on a transparent template brain with the FreeSurfer WM parcellations and whole-brain segmentation. The 11 atlas-based probabilistic tracts from the Mori atlas is shown as 3D renderings, displayed on a semi-transparent template brain from FreeSurfer (fsaverage). The subcortical structures are results from FreeSurfer's whole brain segmentation procedure. The colors on the cortical surface are the cortical parcellations on which the WM parcellations are based (see Figs. 1 and 2). The figure was made by use of 3D Slicer software. |
3D modeling-based surgical planning in transsphenoidal pituitary surgery--preliminary results
|
Publication: Acta Otolaryngol. 2008 Sep;128(9):1011-8. PMID: 19086197 Authors: Raappana A, Koivukangas J, Pirilä T. Institution: Department of Otorhinolaryngology, Head and Neck Surgery, Oulu University Hospital, Oulu, Finland. Background/Purpose: The preoperative three-dimensional (3D) modeling of the pituitary adenoma together with pituitary gland, optic nerves, carotid arteries, and the sphenoid sinuses was adopted for routine use in our institution for all pituitary surgery patients. It gave the surgeon a more profound orientation to the individual surgical field compared with the use of conventional 2D images only. To demonstrate the feasibility of 3D surgical planning for pituitary adenoma surgery using readily available resources. The computed tomography (CT) and magnetic resonance imaging (MRI) data of 40 consecutive patients with pituitary adenoma were used to construct 3D models to be used in preoperative planning and during the surgery. 3D Slicer, a freely available, open source program downloaded to a conventional personal computer (PC) was applied. The authors present a brief description of the 3D reconstruction-based surgical planning workflow. In addition to the preoperative planning the 3D model was used as a 'road map' during the operation. With the 3D model the surgeon was more confident when opening the sellar wall and when evacuating the tumor from areas in contact with vital structures than when using only conventional 2D images. |
A Generic Framework for Internet-based Interactive Applications of High-resolution 3-D Medical Image Data
|
Publication: IEEE Trans Inf Technol Biomed. 2008 Sep;12(5):618-26. PMID: 18779076 Authors: Liu D, Hua KA, Sugaya K. Institution: School of Electrical Engineering and Computer Science, University of Central Florida, Orlando, FL 32816, USA. Background/Purpose: With the advances in medical imaging devices, large volumes of high-resolution 3-D medical image data have been produced. These high-resolution 3-D data are very large in size, and severely stress storage systems and networks. Most existing Internet-based 3-D medical image interactive applications therefore deal with only low- or medium-resolution image data. While it is possible to download the whole 3-D high-resolution image data from the server and perform the image visualization and analysis at the client site, such an alternative is infeasible when the high-resolution data are very large, and many users concurrently access the server. In this paper, we propose a novel framework for Internet-based interactive applications of high-resolution 3-D medical image data. Specifically, we first partition the whole 3-D data into buckets, remove the duplicate buckets, and then, compress each bucket separately. We also propose an index structure for these buckets to efficiently support typical queries such as 3D Slicer and region of interest, and only the relevant buckets are transmitted instead of the whole high-resolution 3-D medical image data. Furthermore, in order to better support concurrent accesses and to improve the average response time, we also propose techniques for efficient query processing, incremental transmission, and client sharing. Our experimental study in simulated and realistic environments indicates that the proposed framework can significantly reduce storage and communication requirements, and can enable real-time interaction with remote high-resolution 3-D medical image data for many concurrent users. |
Anterior cingulate cortex volume reduction in patients with panic disorder
|
Publication: Psychiatry Clin Neurosci. 2008 Jun;62(3):322-30. PMID: 18588593 | PDF Authors: Asami T, Hayano F, Nakamura M, Yamasue H, Uehara K, Otsuka T, Roppongi T, Nihashi N, Inoue T, Hirayasu Y. Institution: Department of Psychiatry, Yokohama City University School of Medicine, Yokohama, Japan. Background/Purpose: Recent neuroimaging studies have suggested that the anterior cingulate cortex (ACC) has an important role in the pathology of panic disorder. Despite numerous functional neuroimaging studies that have elucidated the strong relationship between functional abnormalities of the ACC and panic disorder and its symptoms and response to emotional tasks associated with panic disorder, there has been no study showing volumetric changes of the ACC or its subregions. To clarify the structural abnormalities of ACC and its subregions, the combination of region of interest (ROI) and optimized voxel-based morphometry (VBM) methods were performed on 26 patients with panic disorder, and 26 age and sex-matched healthy subjects. In the ROI study, ACC was divided into four subregions: dorsal, rostral, subcallosal and subgenual ACC. The results of the manually traced ROI volume comparison showed significant volume reduction in the right dorsal ACC. VBM also showed a volume reduction in the right dorsal as well as a part of the rostral ACC as a compound mass. Both manual ROI tracing and optimized VBM suggest a subregion-specific pattern of ACC volume deficit in panic disorder. In addition to functional abnormalities, these results suggest that structural abnormalities of the ACC contribute to the pathophysiology of panic disorder. Funding:
|
A computer modelling tool for comparing novel ICD electrode orientations in children and adults
|
Publication: Heart Rhythm. 2008 Apr;5(4):565-572. PMID: 18362024 | PDF Authors: Matthew Jolley, Jeroen Stinstra, Steve Pieper, Rob Macleod, Dana H. Brooks, Frank Cecchin, John K. Triedman Institution: Department of Cardiology, Children’s Hospital Boston, Boston, MA, USA. Background/Purpose: Use of implantable cardiac defibrillators (ICDs) in children and patients with congenital heart disease is complicated by body size and anatomy. A variety of creative implantation techniques has been used empirically in these groups on an ad hoc basis. OBJECTIVE: To rationalize ICD placement in special populations, we used subject-specific, image-based finite element models (FEMs) to compare electric fields and expected defibrillation thresholds (DFTs) using standard and novel electrode configurations. FEMs were created by segmenting normal torso computed tomography scans of subjects ages 2, 10, and 29 years and 1 adult with congenital heart disease into tissue compartments, meshing, and assigning tissue conductivities. The FEMs were modified by interactive placement of ICD electrode models in clinically relevant electrode configurations, and metrics of relative defibrillation safety and efficacy were calculated. Predicted DFTs for standard transvenous configurations were comparable with published results. Although transvenous systems generally predicted lower DFTs, a variety of extracardiac orientations were also predicted to be comparably effective in children and adults. Significant trend effects on DFTs were associated with body size and electrode length. In many situations, small alterations in electrode placement and patient anatomy resulted in significant variation of predicted DFT. We also show patient-specific use of this technique for optimization of electrode placement. Image-based FEMs allow predictive modeling of defibrillation scenarios and predict large changes in DFTs with clinically relevant variations of electrode placement. Extracardiac ICDs are predicted to be effective in both children and adults. This approach may aid both ICD development and patient-specific optimization of electrode placement. Further development and validation are needed for clinical or industrial utilization. Funding:
|
Using magnetic resonance microscopy to study the growth dynamics of a glioma spheroid in collagen I: A case study
|
Publication: BMC Med Imaging. 2008 Jan 29;8:3. PMID: 18230155 | PDF
Institution: Harvard-MIT (HST) Athinoula A, Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, USA. Background/Purpose: Highly malignant gliomas are characterized by rapid growth, extensive local tissue infiltration and the resulting overall dismal clinical outcome. Gaining any additional insights into the complex interaction between this aggressive brain tumor and its microenvironment is therefore critical. Currently, the standard imaging modalities to investigate the crucial interface between tumor growth and invasion in vitro are light and confocal laser scanning microscopy. While immensely useful in cell culture, integrating these modalities with this cancer's clinical imaging method of choice, i.e. MRI, is a non-trivial endeavour. However, this integration is necessary, should advanced computational modeling be able to utilize these in vitro data to eventually predict growth behaviour in vivo. We therefore argue that employing the same imaging modality for both the experimental setting and the clinical situation it represents should have significant value from a data integration perspective. In this case study, we have investigated the feasibility of using a specific form of MRI, i.e. magnetic resonance microscopy or MRM, to study the expansion dynamics of a multicellular tumor spheroid in a collagen type I gel. |
 MRM images that show the MTS growth over time from different angles. The images to the left depict the axial view, whereas those to the right show an arbitrary angle. These 2D images confirm the surface heterogeneity that emerges already 12 hrs post placement of the MTS into the gel, and depict a small group of invasive cells (images on the right) that can also be seen in Figures 2(A) and 3. On the other hand, images on the left demonstrate that certain parts of the solid spheroid actually grew into the gel (red circle). |
Lowering the Barriers Inherent in Translating Advances in Neuroimage Analysis to Clinical Research Applications
|
Publication: Acad Radiol. 2008 Jan;15(1):114-8. PMID: 18078914 | PDF Authors: Pujol S, Kikinis R, Gollub R. Institution: Department of Radiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA. Background/Purpose: This article presents an initiative for the translation of advances in neuroimage analysis techniques to clinical research scientists. Our objective is to bridge the gap between scientific advances made by the biomedical imaging community and their widespread use in the clinical research community. Through national collaborative effort supported by the National Institutes of Health Roadmap, the integration of the most sophisticated algorithms into usable working open-source systems enables clinical researchers to have access to a broad spectrum of cutting edge analysis techniques. A critical step to maximize the long-term positive impact of this collaborative effort is to translate these techniques into new skills of clinical researchers. To address this challenge, we developed a methodology based on three criteria: a multidisciplinary approach, a balance between theory and common practice, and an immersive collaborative environment. The article illustrates our initiative through the exemplar case of diffusion tensor imaging tractography, and reports on our experience over the past two years of designing and delivering training workshops to more than 300 clinicians and scientists using the developed methodology. Funding:
|
 Exemplar case for Diffusion Tensor Imaging Tractography. Upper left panel displays fiber tractography combined with sagittal and axial slices from a fractional anisotropy map computed with 3D Slicer. Upper-right panel shows fiber tracts generated from regions of interest in the posterior-temporal lobe (yellow), splenium of the corpus callosum (pink), and temporal stem (green). Lower-right figure shows 3D visualization of the computed trajectory of the inferior occipito-frontal fasciculus bundle (red). |
2007
|
Publication: Comput Aided Surg. 2007 Nov;12(6):311-24. PMID: 18066947 | PDF Authors: Raúl San José Estépar, Nicholas Stylopoulos, Randy Ellis, Eigil Samset, Carl-Fredrik Westin, Christopher Thompson, Kirby Vosburgh Institution: Department of Radiology, Harvard Medical School and Brigham and Women's Hospital, Boston, MA, USA. Background/Purpose: Scarless surgery is an innovative and promising technique that may herald a new era in surgical procedures. We have created a navigation system, named IRGUS, for endoscopic and transgastric access interventions and have validated it in in vivo pilot studies. Our hypothesis is that endoscopic ultrasound procedures will be performed more easily and efficiently if the operator is provided with approximately registered 3D and 2D processed CT images in real time that correspond to the probe position and ultrasound image. The system provides augmented visual feedback and additional contextual information to assist the operator. It establishes correspondence between the real-time endoscopic ultrasound image and a preoperative CT volume registered using electromagnetic tracking of the endoscopic ultrasound probe position. Based on this positional information, the CT volume is reformatted in approximately the same coordinate frame as the ultrasound image and displayed to the operator. The system reduces the mental burden of probe navigation and enhances the operator's ability to interpret the ultrasound image. Using an initial rigid body registration, we measured the mis-registration error between the ultrasound image and the reformatted CT plane to be less than 5 mm, which is sufficient to enable the performance of novice users of endoscopic systems to approach that of expert users. Our analysis shows that real-time display of data using rigid registration is sufficiently accurate to assist surgeons in performing endoscopic abdominal procedures. By using preoperative data to provide context and support for image interpretation and real-time imaging for targeting, it appears probable that both preoperative and intraoperative data may be used to improve operator performance. Funding:
|
3D visualization and simulation of frontoorbital advancement in metopic synostosis
|
Publication: Childs Nerv Syst. 2007 Nov;23(11):1313-7. PMID: 17701413 | PDF Authors: Rodt T, Schlesinger A, Schramm A, Diensthuber M, Rittierodt M, Krauss JK. Institution: Department of Radiology, Hannover University Medical School, Carl-Neuberg-Str. 1, 30625 Hannover, Germany. Background/Purpose: Current multislice computed tomography (CT) technology can be used for diagnosis and surgical planning applying computer-assisted three-dimensional (3D) visualization and surgical simulation. The usefulness of a technique for surgical simulation of frontoorbital advancement is demonstrated here in a child with metopic synostosis. Postprocessing of multi-slice CT data was performed using the software 3D Slicer. 3D models were created for the purpose of surgical simulation. These allow planning the course of the osteotomies and individually placing the different bony fragments by an assigned matrix to simulate the surgical result. Photo documentation was obtained before and after surgery. Surgical simulation of the procedure allowed determination of the osteotomy course and assessment of the positioning of the individual bony fragments. Computer-assisted postprocessing and simulation is a useful tool for surgical planning in craniosynostosis surgery. The time-effort for segmentation currently limits the routine clinical use of this technique. |
Creating physical 3D stereolithograph models of brain and skull]
|
Publication: PLoS One. 2007 Oct; 2(10): e1119. PMID: 17971879 | PDF Authors: Kelley DJ, Farhoud M, Meyerand ME, Nelson DL, Ramirez LF, Dempsey RJ, Wolf AJ, Alexander AL, Davidson RJ. Institution: Waisman Laboratory for Brain Imaging and Behavior, Waisman Center, Madison, WI, USA. Background/Purpose: The human brain and skull are three dimensional (3D) anatomical structures with complex surfaces. However, medical images are often two dimensional (2D) and provide incomplete visualization of structural morphology. To overcome this loss in dimension, we developed and validated a freely available, semi-automated pathway to build 3D virtual reality (VR) and hand-held, stereolithograph models. To evaluate whether surface visualization in 3D was more informative than in 2D, undergraduate students (n = 50) used the Gillespie scale to rate 3D VR and physical models of both a living patient-volunteer's brain and the skull of Phineas Gage, a historically famous railroad worker whose misfortune with a projectile tamping iron provided the first evidence of a structure-function relationship in brain. Using our processing pathway, we successfully fabricated human brain and skull replicas and validated that the stereolithograph model preserved the scale of the VR model. Based on the Gillespie ratings, students indicated that the biological utility and quality of visual information at the surface of VR and stereolithograph models were greater than the 2D images from which they were derived. The method we developed is useful to create VR and stereolithograph 3D models from medical images and can be used to model hard or soft tissue in living or preserved specimens. Compared to 2D images, VR and stereolithograph models provide an extra dimension that enhances both the quality of visual information and utility of surface visualization in neuroscience and medicine. Funding:
|
 Stereolithograph of Phineas Gage's Skull. The processing pathway we developed can replicate preserved specimens. (a) Virtual model of Phineas Gage's skull. (b) With this physical 3D model of Phineas Gage's skull, we illustrate the approximate path of the tamping iron that produced Phineas Gage's famous injury. |
Morphology, constraints, and scaling of frontal sinuses in the hartebeest, Alcelaphus buselaphus (Mammalia: Artiodactlya, Bovidae)
|
Publication: Journal of Morphology 2007; 268:243-253. PMID: 17278134 | PDF. See also 3D Slicer: The Tutorial Part V. Authors: Farke, A. A. Institution: Department of Anatomical Sciences, Stony Brook University, N.Y. Background/Purpose: The frontal sinuses of bovid mammals display a great deal of diversity, which has been attributed to both phylogenetic and functional influences. In-depth study of the hartebeest (Alcelaphus buselaphus), a large African antelope, reveals a number of previously undescribed details of frontal sinus morphology. In A. buselaphus, the frontal sinuses conform closely to the shape of the frontal bone, filling nearly the entire element. However, the horncores are never extensively pneumatized, contrasting with the condition seen in many other bovids. This evidence is inconsistent with the hypothesis that sinuses are opportunistic pneumatizing agents, suggesting that phylogenetic factors also play a role in determining sinus size. Both cranial sutures and neurovasculature appear to constrain the growth of sinuses in part. In turn, the sinus also affects the growth of the parietal; apparently this element is not truly pneumatized by the sinus in most cases, but the bone’s shape changes under the influence of the sinus. Furthermore, the sinuses present relatively few struts when compared with the sinuses of some other bovids, such as Ovis. By adapting methods previously developed for measuring structural parameters of trabecular bone, it is possible to quantify certain aspects of sinus morphology. These include the number of bony struts within the sinus, the spacing of these struts, and the size of individual cavities within the sinus. Some differences in the number of struts are evident between subspecies. Similarly, significant differences occur in the relative number of struts between male and female A. buselaphus, which may be related to behavior. The volume of the sinus is strongly correlated with the size of the frontal, but less so with overall cranial size. This finding illustrates the importance of choosing variables carefully when comparing sinus sizes and growth between species. |
Image-guided Otologic Surgery based on Patient Motion Compensation and Intraoperative Virtual CT
|
Publication: Proceedings of ACCAS2007, #P110064 Authors: Hong J, Matsumoto N, Ouchida R, Komune S, Hashizume M Institution: Redox Navi Institute, Kyushu University, Japan. Background/Purpose: Image-guided surgery has been established particularly in ear, nose, throat (ENT) surgery that requires precise approach in small and complicated anatomy. During the surgery, the registration between the patient and the radiographic images must be maintained regardless of the patient movement. In this study, relative position and orientation of the surgical instrument is calculated referring to the moving patient position. Since the system does not directly handle the 3-D volume data, real-time update of 3-D navigational view is possible. The proposed method also keeps recording the surgical drill position while tracking it. With these digitized data, the temporal bone resection area is computed with several rule-based image processing. Finally, virtual CT images are constructed, and presented for the surgeon to understand and evaluate the current operation intraoperatively by observing their familiar CT images. The aim of this study is to develop and evaluate a novel otologic navigation system supporting patient motion compensation and intraoperative virtual CT construction. |
Non-rigid registration of pre-procedural MR images with intra- procedural unenhanced CT images for improved targeting of tumors during liver radiofrequency ablations
|
Publication: Int Conf Med Image Comput Comput Assist Interv. 2007;10(Pt 2):969-77. PMID: 18044662 | PDF Authors: N Archip, S Tatli, PR Morrison, F Jolesz, SK Warfield, SG Silverman Institution: Department of Radiology, Harvard Medical School and Brigham and Women's Hospital, Boston, MA, USA. Background/Purpose: In the United States, unenhanced CT is currently the most common imaging modality used to guide percutaneous biopsy and tumor ablation. The majority of liver tumors such as hepatocellular carcinomas are visible on contrast-enhanced CT or MRI obtained prior to the procedure. Yet, these tumors may not be visible or may have poor margin conspicuity on unenhanced CT images acquired during the procedure. Non-rigid registration has been used to align images accurately, even in the presence of organ motion. However, to date, it has not been used clinically for radiofrequency ablation (RFA), since it requires significant computational infrastructure and often these methods are not sufficient robust. We have already introduced a novel finite element based method (FEM) that is demonstrated to achieve good accuracy and robustness for the problem of brain shift in neurosurgery. In this current study, we adapt it to fuse pre-procedural MRI with intra-procedural CT of liver. We also compare its performance with conventional rigid registration and two non-rigid registration methods: b-spline and demons on 13 retrospective datasets from patients that underwent RFA at our institution. FEM non-rigid registration technique was significantly better than rigid (p<10-5), non-rigid b-spline (p<10- 4) and demons (p<10-4) registration techniques. The results of our study indicate that this novel technology may be used to optimize placement of RF applicator during CT-guided ablations. Funding:
|
A detailed 3D model of the guinea pig cochlea
|
Publication: Brain Struct Funct. 2007 Sep;212(2):223-30. PMID: 17717692 Authors: Liu B, Gao XL, Yin HX, Luo SQ, Lu J. Institution: Department of Anatomy, Capital Medical University, 10#, You An Men Wai, Xi Tou Tiao, Beijing 100069, China. Background/Purpose: Several partial models of cochlear subparts are available. However, a complete 3D model of an intact cochlea based on actual histological sections has not been reported. Hence, the aim of this study was to develop a novel 3D model of the guinea pig cochlea and conduct post-processes on this reconstructed model. We used a combination of histochemical processing and the method of acquiring section data from the visible human project (VHP) to obtain a set of ideal raw images of cochlear sections. After semi-automatic registration and accurate manual segmentation with professional image processing software, one set of aligned data and six sets of segmented data were generated. Finally, the segmented structures were reconstructed by 3D Slicer (a professional imaging process and analysis tool). Further, post-processes including 3D visualization and a virtual endoscope were completed to improve visualization and simulate the course of the cochlear implant through the scala tympani. The 3D cochlea model contains the main six structures: (1) the inner wall, (2) modiolus and spiral lamina, (3) cochlea nerve and spiral ganglion, (4) spiral ligament and inferior wall of cochlear duct, (5) Reissner's membrane and (6) tectorial membrane. Based on the results, we concluded that ideal raw images of cochlear sections can be acquired by combining the processes of conventional histochemistry and photographing while slicing. After several vital image processing and analysis steps, this could further generate a vivid 3D model of the intact cochlea complete with internal details. This novel 3D model has great potential in teaching, basic medical research and in several clinical applications. |
Dynamic 3-D computer modeling tracks brain changes during surgery
|
Publication: National Science Foundation (NSF) Discoveries - Brain Surgery: It Really Is Brain Surgery, August 31, 2007. Authors: Nikos Chrisochoides Institution: College of William and Mary Background/Purpose: In essence, the William and Mary team provides the surgical team with a dynamic computer model of the patient's brain. In clinical trials, Chrisochoides says his team can render a new model in six or seven minutes, but hopes to be able to do so in under two minutes. More... Funding:
|
Surface Rendering-based Virtual Intraventricular Endoscopy: Retrospective Feasibility Study and Comparison to Volume Rendering-based Approach
|
Publication: Neuroimage. 2007;37 Suppl 1:S89-99. PMID: 17513130 | PDF Authors: Nakajima N, Wada J, Miki T, Haraoka J, Hata N. Institution: Department of Radiology, Brigham and Women's Hospital, Boston, MA, USA. Background/Purpose: Virtual endoscopic simulations using volume rendering (VR) have been proposed as a tool for training and understanding intraventricular anatomy. It is not known whether surface rendering (SR), an alternative to VR, can visualize intraventricular and subependymal structures better and thus making the virtual endoscope more useful for simulating the intraventicular endoscopy. We sought to develop SR virtual endsocopy and compared the visibility of anatomical structures in SR and VR using retrospective cases. Fourteen patients who underwent endoscopic intraventricular surgery of third ventricle enrolled the study. SR virtual endoscopy module was developed in open-source software 3D Slicer and virtual endoscopic scenes from the retrospective cases were created. VR virtual endoscopy of the same cases was prepared in commercial software. Three neurosurgeons scored the visibility of substructures in lateral and third ventricle, arteries, cranial nerves, and other lesions. We found that VR and SR virtual endoscopy performed similarly in visualization of substructures in lateral and third ventricle (not significant statistically). However, the SR was statistically significantly better in visualizing subependymal arteries, cranial nerves, and other lesions (p<0.05, respectively). Funding:
|
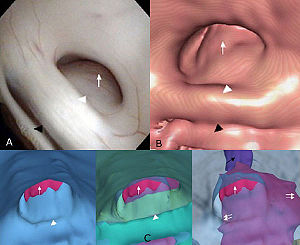 Visualization of the subependymal extension of the tumor in endoscopic surgical simulation using 3D Slicer. (A) The real intraoperative image (flexible videoscope) presents the cerebral aqueduct obstructed by the tectal tumor. (B) Volume rendering-based virtual endoscopy provides the similar image to the real intraoperative image. (C) Surface renderingbased virtual endoscopy reveals the subependymal extension of the tumor by changing the opacity and visibility of the other structures. Tumor (white arrow); posterior commissure (white arrowhead); habenular commissure (black arrowhead); cerebral aqueduct behind the tumor (black arrow). |
Interest of the Steady State Free Precession (SSFP) sequence for 3D modeling of the whole fetus
|
Publication: Proceedings of the 29th Annual International Conference of the IEEE EMBS, 2007. | PDF Authors: Anquez, J., Angelini, E., Bloch, I., Merzoug, V., Bellaiche-Millischer, A.E., Adamsbaum, C. Institution: Signal and Image Processing Department, GET - Telecom Paris, France. Background/Purpose: Fetal magnetic resonance imaging (MRI) has been gaining interest over the last two decades. Current fast MRI sequences provide imaging data of the whole uterus in less than 20 seconds, avoiding fetal motion related artifacts without any maternal or fetal sedation. MRI has proved to be a useful adjunct to echographic screening for prenatal diagnosis. However, MRI volumetric data is still mainly interpreted on 2D slices and 3D applications remain limited. In this paper, we discuss the qualities of the SSFP MRI sequences to provide adequate data for 3D segmentation and modeling of the fetus. Potential exploitations of 3D segmentation and derived anatomical models cover several domains: biometric and morphologic clinical studies, quantitative longitudinal studies of normal and abnormal fetus developments, direct visualization of the overall fetus body and simulations in different fields (surgery, radiation dosimetry,...). |
Longitudinal in vivo Reproducibility of Cartilage Volume and Surface in Osteoarthritis of the Knee
|
Publication: Skeletal Radiol. 2007 Apr;36(4):315-20. PMID: 17219231 | PDF Authors: Brem MH, Pauser J, Yoshioka H, Brenning A, Stratmann J, Hennig FF, Kikinis R, Duryea J, Winalski CS, Lang P. Institution: Musculoskeletal Division, Department of Radiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA. Background/Purpose: The aim of this study was to evaluate the longitudinal reproducibility of cartilage volume and surface area measurements in moderate osteoarthritis (OA) of the knee. We analysed 5 MRI (GE 1.5T, sagittal 3D SPGR) data sets of patients with osteoarthritis (OA) of the knee (Kellgren Lawrence grade I-II). Two scans were performed: one baseline scan and one follow-up scan 3 months later (96±10 days). For segmentation, 3D Slicer 2.5 software was used. Two segmentations were performed by two readers independently who were blinded to the scan dates. Tibial and femoral cartilage volume and surface were determined. Longitudinal and cross-sectional precision errors were calculated using the standard deviation (SD) and coefficient of variation (CV%=100x[SD/mean]) from the repeated measurements in each patient. The in vivo reproducibility was then calculated as the root mean square of these individual reproducibility errors. The cross-sectional root mean squared coefficient of variation (RMSE-CV) was 1.2, 2.2 and 2.4% for surface area measurements (femur, medial and lateral tibia respectively) and 1.4, 1.8 and 1.3% for the corresponding cartilage volumes. Longitudinal RMSE-CV was 3.3, 3.1 and 3.7% for the surface area measurements (femur, medial and lateral tibia respectively) and 2.3, 3.3 and 2.4% for femur, medial and lateral tibia cartilage volumes. The longitudinal in vivo reproducibility of cartilage surface and volume measurements in the knee using this segmentation method is excellent. To the best of our knowledge we measured, for the first time, the longitudinal reproducibility of cartilage volume and surface area in participants with mild to moderate OA. Funding:
|
Origin and insertion points involved in levator ani muscle defects
|
Publication: Am J Obstet Gynecol. 2007 Mar;196(3):251.e1-5. PMID: 17346542 | PDF Authors: Margulies RU, Huebner M, DeLancey JO. Institution: Pelvic Floor Research Group, University of Michigan, Ann Arbor, MI, USA. Background/Purpose: This project sought to identify and to describe the anatomical connections affected by levator ani defects involving the pubovisceral portion of the muscle. Fourteen magnetic resonance scans of women with unilateral levator defects were selected. The missing muscle mapping technique was used to characterize the absent muscle. Normal muscle was visualized and compared with the contralateral side. Using a 3D Slicer, the outline of the intact muscle was traced; models of this muscle and surrounding structures were generated. The missing muscle originates from the posterior pubic bone and extends laterally over the obturator internus muscle; it inserts into the vaginal wall, perineal body, and the intersphincteric space. Architectural distortion, with an asymmetric lateral spilling of the vagina was present in 50% of women. The defect was right sided in 71% of patients. The origin and insertion points of the damaged portion of the levator ani muscle were identified. Funding:
|
A prototype biosensor-integrated image-guided surgery system
|
Publication: Int J Med Robot. 2007 Mar;3:82-8. PMID: 17441030 | PDF Authors: Reisner LA, King BW, Klein MD, Auner GW, Pandya AK. Institution: Department of Electrical and Computer Engineering, Wayne State University, Detroit, MI, USA. Background/Purpose: In this study we investigated the integration of a Raman spectroscopy-based biosensor with an image-guided surgery system. Such a system would provide a surgeon with both a diagnosis of the tissue being analysed (e.g. cancer) and localization information displayed within an imaging modality of choice. This type of mutual and registered information could lead to faster diagnoses and enable more accurate tissue resections. METHODS: A test bed consisting of a portable Raman probe attached to a passively articulated mechanical arm was used to scan and classify objects within a phantom skull. The prototype system was successfully able to track the Raman probe, classify objects within the phantom skull, and display the classifications on medical imaging data within a virtual reality environment. We discuss the implementation of the integrated system, its accuracy and improvements to the system that will enhance its usefulness and further the field of sensor-based computer-assisted surgery. Funding:
|
|
Publication: Int J CARS (2007) 2:65–73. | PDF Authors: Georgi Diakov, Wolfgang Freysinger Institution: 4D Visualization Laboratory, University Clinic of Oto-, Rhino-, Laryngology, Innsbruck Medical University, Innsbruck, Austria Background/Purpose: An initialization-free approach for perioperative registration in functional endoscopic sinus surgery (FESS) is sought. The quality of surgical navigation relies on registra- tion accuracy of preoperative images to the patient. Although landmark-based registration is fast, it is prone to human oper- ator errors. This study evaluates the accuracy of two well- known methods for segmentation of the occipital bone from CT-images for use in surgical 3D-navigation. Funding:
|
 A zoom in the mesh, displayed in 3D Slicer. The oversampling is seen as three layers at each “step”. The difference between the TMCS mesh and the “ground truth” is seen in the colored spots (“ground truth” green, TMCS pink). The three circles mark zones with visibly discernible differences in the segmentation; the inset is a magnification of one of these zones |
Development of a three-dimensional multiscale agent-based tumor model: simulating gene-protein interaction profiles, cell phenotypes and multicellular patterns in brain cancer
|
Publication: J Theor Biol. 2007 Jan 7;244(1):96-107. PMID: 16949103 | PDF Authors: Zhang L, Athale CA, Deisboeck TS. Institution: Complex Biosystems Modeling Laboratory, Massachusetts General Hospital Background/Purpose: Related also to efforts within the Center for the Development of a Virtual Tumor, CViT (http://www.cvit.org), our research here (http://biosystems.mit.edu) focuses on developing multi-scale and multi-resolution (brain) cancer models. Using a hybrid 3D agent-based modeling platform we connect gene-protein interaction maps on the molecular and single cell level (Figure 1, left) up to the volumetric tumor scale of clinical MR-images. For the latter, we plan on using 3D Slicer for segmentation of tumor-affected areas and 3D reconstruction of tumor volume and surface. Figure 1, right depicts a first step towards this approach where the yellow points depict the 3D surface reconstruction of a T1+Gd MRI while the blue and red points represent virtual tumor cells in attempt to match the real data points. Amongst the more challenging steps is the translation of these 2D in vivo imaging data onto our in silico lattices. Funding:
|
Image Registered FAST (IRFAST) for Combat Casualty Triage
|
Authors: Kilian Pohl in collaboration with Steve Pieper, Kirby Vosburgh, and Barnabas Takacs. Background/Purpose: Dr. Pohl's research involves developing an automatic segmentation approach identifying major anatomical compartments from torso CT scans. Using 3D Slicer, he developed a procedure for automatically segmenting the body, bone structures, lungs, and hart from CT images (see figure). The method iterates between the identification of anatomical structures and the registration of an “atlas” to the CT data set of the subject. The atlas is based on CT data set, which we call template, and a label map that already identifies the structures of interest in the template. In the first iteration, the template with the segmentation of the body is mapped onto the CT data set of the subject. This results in a very accurate segmentation of the subject’s body. The method then combines the segmentation of the body and the CT images of the subject to identify the lungs and bone structures. In the second iteration, the method maps the atlas specific label map of body, bones, and lungs to the corresponding label map of the subject. The segmented structures are then used as a "coordinate system" used for the identification of the remaining anatomical compartments. Funding:
|
A Preliminary Study on the Relationship between Nasal Cavity and Maxillary Sinus Volumes
|
Publication: Proceedings of the 76th Annual Meeting of the American Association of Physical Anthropologists March 2007. | PDF Authors: L.N. Butaric, D.C. Broadfield, R.C. McCarthy Institution: Department of Anthropology, Texas A&M University Background/Purpose: Previous research suggests that nasal cavity volume (NCV), thought to be related to climate, is inversely correlated with maxillary sinus volume (MSV). According to this hypothesis, changes in nasal cavity size and shape reflect physiological needs, such as warming and humidifying inspired air. Owing to volumetric constraints in the mid-face, relative increases in NCV are hypothesized to result in concomitant decreases in MSV, respectively. To test this hypothesis thirty-nine dried adult human crania from seven different climatic regions were examined using computerized tomography (CT) scans. Significant differences in MSV and NCV between populations were identified using Analysis of Variance (ANOVA). In addition, least-squares and reduced major axis (RMA) regression analyses were performed to test the scaling relationships between MSV, NCV and several cranial size variables. Contrary to previous studies, results indicate that MSV and NCV are not significantly correlated. RMA analyses indicate that NCV, but not MSV, scales isometrically with skull size. Finally, post hoc ANOVA results identify significant differences between human populations for MSV that do not follow climatic or environmental trends. These results suggest that (1) it is unlikely that NCV and MSV compete for space in the mid-face, (2) NCV is largely a byproduct of skull size, and (3) NCV and MSV may not be as closely tied to climate as previously thought. Additional genetic and epigenetic factors need to be considered regarding the structure and function of the human maxillary sinus. |
Interobserver variability in the determination of upper lobe-predominant emphysema
|
Publication: Chest. 2007 Feb;131(2):424-31. PMID: 17296643 | PDF Authors: Hersh CP, Washko GR, Jacobson FL, Gill R, Estepar RS, Reilly JJ, Silverman EK. Institution: Channing Laboratory, Brigham and Women's Hospital, 181 Longwood Ave, Boston, MA, USA. Background/Purpose: Appropriateness for lung volume reduction surgery is often determined based on the results of high-resolution CT (HRCT) scanning of the chest. At many centers, radiologists and pulmonary physicians both review the images, but the agreement between readers from these specialties is not known. Two thoracic radiologists and three pulmonologists retrospectively reviewed the HRCT scans of 30 patients with emphysema involved in two clinical studies at our institution. Each reader assigned an emphysema severity score and assessed upper lobe predominance, using a methodology similar to that of the National Emphysema Treatment Trial. In addition, the percentage of emphysema at -910 Hounsfield units was objectively determined by density mask analysis. For the emphysema severity scores, (Spearman) correlation between readers ranged from 0.59 (p = 0.0005) to 0.87 (p < 0.0001), with generally stronger correlations among readers from the same medical specialty. Emphysema severity scores were significantly correlated with prebronchodilator and postbronchodilator spirometry findings, as well as with density mask analysis. In the assessment of upper lobe predominance, kappa statistics for agreement ranged from 0.20 (p = 0.4) to 0.60 (p = 0.0008). Examining all possible radiologist-pulmonologist pairs, the two readers agreed in their assessments of emphysema distribution in 75% of the comparisons. Readers agreed on upper lobe-predominant disease in 9 of the 10 patients in which regional density mask analysis clearly showed upper lobe predominance. In a group of patients with varying emphysema severity, interobserver agreement in the determination of upper lobe-predominant disease was poor. Agreement between readers tended to be better in cases with clear upper lobe predominance as determined by densitometry. Funding:
|
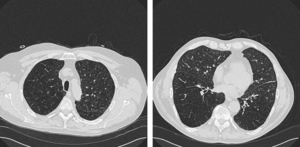 Example of disagreement among readers. Two of five observers interpreted this HRCT scan as showing upper lobe-predominant emphysema. Representative 1-mm sections from the level of (left, A) the top of the aortic arch and (right, B) the right inferior pulmonary vein are shown, using lung windows (width, –1,500 HU; level, 600 HU). |
Registered, Sensor-Integrated Virtual Reality for Surgical Applications
|
Publication: Proceedings of the International Conference and Exhibition in Virtual Reality 2007; 277-278. | PDF Authors: Brady W. King, Luke A. Reisner, Michael D. Klein, Gregory W. Auner, Abhilash K. Pandya Institution: Wayne State University, Detroit, Michigan, USA Background/Purpose: The visualization for our image-guided surgery system is implemented using 3D Slicer, an open-source application for displaying medical data. 3D Slicer provides a virtual reality environment in which various imaging modalities (e.g. CT or MRI data) can be presented. The software includes the ability to display the locations of objects with respect to 3D models that are derived from segmentation of the medical imaging. We modified 3D Slicer in several ways to adapt it to our application. First, we developed a TCP/IP interface that receives the tracking data for the MicroScribe and displays its position in the VR environment relative to the medical imaging data. This allows us to track the Raman probe in real-time. Second, we developed a way to place colored markers that indicate tissue/material classification on the medical imaging data. The combination of these modifications enables us to denote the location and classification of tissue/material scanned with the probe in near-real-time. |
Modeling Cancer Biology
|
Publication: Biomedical Computation Review Spring 2007 pp. 16-24. | PDF Author: Kristin Cobb |
A Comparison of Upper Airway Structures in Male and Female Obstructive Sleep Apnoea (OSA) Patients
|
Publication: Poster Presentation at the 2007 American Thoracic Society International Conference Authors: RD McEvoy, MK Ryan, PG Catcheside, M Jakab, A Malhotra, MR Sage, R Valentine, DP White Institution: Adelaide Institute for Sleep Health, Repatriation General Hospital, SA Australia. Background/Purpose: For the same BMI female OSA patients tend to have less severe sleep disordered breathing than male OSA patients. This study investigates features of upper airway anatomy that may contribute to this difference. Male (n=6) and female (n=9) patients matched for age [mean(SD); men 56.5(5.7), women 55.3(6.2) yrs] and BMI [35.2(3.4), 34.3 (3.0) kg/m²], who had previously been diagnosed with OSA (AHI>30/h), and adherent with CPAP for >3 months were studied. T1 weighted magnetic resonance imaging of the upper airway and surrounding tissues was performed and analyzed (3D Slicer) to determine airway, soft tissue and bony dimensions. The oropharyngeal bony vault volume was greater in males after normalizing for height (1.2(0.1) vs 0.97(0.1), mls/cm, p=0.001). Men had greater neck circumference (45.3(4.3) vs 39.5(4.5) cm, p=0.026) and longer pharyngeal airways (75.2(8.3) vs 64.6(9.5) mm, p=0.042) compared to women. Pharyngeal airway volume and minimum CSA were not different between the genders. In the mid sagittal plane, the tongue CSA was greater in men before and after (36.4(1.4) vs 32.6(3.5) mm²/mm, p=0.01) normalizing for bony vault dimensions. In data collected to date, AHI was not different between males and females (55(12) vs 62(21) events/h). These preliminary data suggest that male OSA patients have greater oropharyngeal bony vault volume than female patients; a larger tongue (normalized for bony vault dimensions) and a longer pharyngeal airway. These findings may have implications for the male predisposition for OSA. Funding:
|
2006
Shaving diffusion tensor images in discriminant analysis: a study into schizophrenia
|
Publication: Med Image Anal. 2006 Dec;10(6):841-9. PMID: 16965928 | PDF Authors: Caan MW, Vermeer KA, van Vliet LJ, Majoie CB, Peters BD, den Heeten GJ, Vos FM. Institution: Quantitative Imaging Group, Delft University of Technology, Delft, The Netherlands. Background/Purpose: A technique called 'shaving' is introduced to automatically extract the combination of relevant image regions in a comparative study. No hypothesis is needed, as in conventional pre-defined or expert selected region of interest (ROI)-analysis. In contrast to traditional voxel based analysis (VBA), correlations within the data can be modeled using principal component analysis (PCA) and linear discriminant analysis (LDA). A study into schizophrenia using diffusion tensor imaging (DTI) serves as an application. Conventional VBA found a decreased fractional anisotropy (FA) in a part of the genu of the corpus callosum and an increased FA in larger parts of white matter. The proposed method reproduced the decrease in FA in the corpus callosum and found an increase in the posterior limb of the internal capsule and uncinate fasciculus. A correlation between the decrease in the corpus callosum and the increase in the uncinate fasciculus was demonstrated. Funding:
|
|
Publication: Med Bio Eng Comput. 2006 Dec; 44(12):1127-1134. PMID: 17102954 Authors: Hong J, Nakashima H, Konishi K, Ieiri S, Tanoue K, Hashizume M Institution: Redox Navi Institute, Kyushu University, Japan. Background/Purpose: Surgical navigation systems can contribute to abdominal interventions such as radiofrequency ablation (RFA) or percutaneous ethanol injection therapy (PEIT) for liver tumours. Until now, typical abdominal interventions have been performed using the US guidance approach. Identification of the lesion by these methods has often been difficult, especially when tumours were small and US images were unclear. To improve the treatment with better lesion identification, 3D navigation based on MRI or CT is crucial. Particularly, MRI-based navigation systems have been preferred in respect of avoidance of ionizing radiation. To our knowledge, this is the first trial of abdominal needle insertion intervention under local anaesthesia using simultaneous MRI and US navigation. To accomplish this purpose, the semi-automatic fiducial registration is required. While scanning the abdominal area and using the optical tracking system, partial loss of the markers often occurs, leading to the prolongation of the limited intervention time. The proposed method combats this problem. In this paper, we elucidate whether the MRI and US integrated navigation assists in reliable needle insertion and reduce the time required to determine the needle insertion path in abdominal interventions. |
Molar crown thickness, volume, and development in South African Middle Stone Age humans
|
Publication: South African Journal of Science. 2006;102(11-12):513-517. | PDF Authors: SMITH Tanya M., OLEJNICZAK Anthony J., TAFFOREAU Paul, REID Donald J., GRINE Fredrick E., HUBLIN Jean-Jacques Institution: Department of Human Evolution, Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany. Background/Purpose: One highly debated issue in palaeoanthropology is that of modern human origins, particularly the issue of when 'anatomically modern humans' (AMH) from the African Middle Stone Age became fully modern. While studies of cranial and external dental morphology suggest a modern transition occurred 150 000-200 000 years ago, little is known about dental development or enamel thickness in AMH. Studies of early members of the genus Homo suggest that the modern, prolonged condition of tooth growth arose late in human evolution, and that the enamel thickness of earlier hominins may not be homologous to the modern condition. This study represents the first integrated investigation of molar crown enamel thickness, volume, and development in fossil hominins, aimed at determining whether differences between AMH and living populations can be detected in these traits. Using high-resolution micro-computed tomography, we demonstrate similarities in enamel thickness and crown volumes between fossil and modern populations. Additionally, long-period growth line numbers and estimates of crown formation times for AMH molars fall within modern human ranges. These findings suggest that tooth structure and growth have remained constant for more than 60 000 years, despite the known geographical, technological, and ecological diversity that characterizes later stages of human evolution. |
 Virtually separated coronal dentine (left) and enamel cap (right) of the right lower first molar illustrated in Figs 1 and 3. A small amount of enamel is missing on the lingual cervix. The volume and surface area of each of these tissues was calculated, which was used to calculate the 3D average and relative enamel thickness (see text). |
A 3D Model Simulating Sediment Transport, Erosion and Deposition within a Network of Channel Belts and an Associated Floodplain
|
Publication: PowerPoint Presentation PDF Authors: Derek Karssenberg and John Bridge Institution: Utrecht University, the Netherlands and Binghamton University, NY, USA. |
Molecular Diffusion in MRI: Technical Application of Fiber Tracking
|
Publication: PowerPoint Presentation PPT Author: Martha Liliana Mora V. Institution: Rey Juan Carlos University, Madrid, Spain. |
Quantification of Levator Ani Cross-sectional Area Differences between Women with and those without Prolapse
|
Publication: Obstet Gynecol. 2006 Oct;108(4):879-83. PMID: 17012449 | PDF Authors: Hsu Y, Chen L, Huebner M, Ashton-Miller JA, DeLancey JO. Institution: University of Michigan, Ann Arbor, Michigan, USA Background/Purpose: Compare levator ani cross-sectional area as a function of prolapse and muscle defect status. Thirty women with prolapse and 30 women with normal pelvic support were selected from an ongoing case-control study of prolapse. For each of the two groups, 10 women were selected from three categories of levator defect severity: none, minor, and major identified on supine magnetic resonance scans. Using those scans, three-dimensional (3D) models of the levator ani muscles were made using a modeling program (3D Slicer), and cross-sections of the pubic portion were calculated perpendicular to the muscle fiber direction using another program, I-DEAS. An analysis of variance was performed. The ventral component of the levator muscle of women with major defects had a 36% smaller cross-sectional area, and women with minor defects had a 29% smaller cross-sectional area compared with the women with no defects (P < .001). In the dorsal component, there were significant differences in cross-sectional area according to defect status (P = .03); women with major levator defects had the largest cross-sectional area compared with the other defect groups. For each defect severity category (none, minor, major), there were no significant differences in cross-sectional area between women with and those without prolapse. Women with visible levator ani defects on magnetic resonance imaging had significantly smaller cross-sectional areas in the ventral component of the pubic portion of the muscle compared with women with intact muscles. Women with major levator ani defects had larger cross-sectional areas in the dorsal component than women with minor or no defects. Funding:
|
Liver metastases: 3D shape-based analysis of CT scans for detection of local recurrence after radiofrequency ablation
|
Publication: Radiology. 2006 Oct;241(1):243-50. PMID: 16928977 | PDF Authors: Ivan Bricault, Ron Kikinis, Paul R Morrison, Eric Vansonnenberg, Kemal Tuncali, Stuart G Silverman Institution: Department of Radiology, University Hospital Michallon, Grenoble, France Background/Purpose: The purpose of this pilot study was to determine retrospectively the diagnostic performance of a computer-aided three-dimensional (3D) analytic tool for assessing local recurrences of liver metastases by quantifying shape changes in ablated tumors on computed tomographic (CT) scans for follow-up of radiofrequency (RF) ablation. Positron emission tomographic and long-term CT follow-up images were reference standards. Fifty-six follow-up CT scans of 12 liver metastases (mean size, 4.0 cm) in nine patients treated with RF ablation were retrospectively analyzed. After the 1st month following RF ablation, the 3D analytic tool helped quantify ablated tumor shape variations and revealed recurrences even in the absence of abnormal enhancement (sensitivity, seven of seven; specificity, three of five). The 3D tool would have revealed a recurrence before it was reported clinically in two patients. Although results are preliminary, a 3D analytic tool based on shape may be useful in assessing RF ablation results. Funding:
|
Atlas Guided Identification of Brain Structures by Combining 3D Segmentation and SVM Classification
|
Publication: Int Conf Med Image Comput Comput Assist Interv. 2006 Oct;9(Pt 2):209-16. PMID: 17354774 | PDF Authors: Ayelet Akselrod-Ballin, Meirav Galun, Moshe John Gomori, Ronen Basri, and Achi Brandt Institution: Department of Computer Science and Applied Math, Weizmann Institute of Science, Rehovot, Israel and Department of Radiology, Hadassah University Hospital, Jerusalem, Israel Background/Purpose: This study presents a novel automatic approach for the identification of anatomical brain structures in magnetic resonance images (MRI). The method combines a fast multiscale multi-channel three dimensional (3D) segmentation algorithm providing a rich feature vocab ulary together with a support vector machine (SVM) based classifier. The segmentation produces a full hierarchy of segments, expressed by an irregular pyramid with only linear time complexity. The pyramid provides a rich, adaptive representation of the image, enabling detection of various anatomical structures at different scales. A key aspect of the approach is the thorough set of multiscale measures employed throughout the segmentation process which are also provided at its end for clinical analysis. These features include in particular the prior probability knowl- edge of anatomic structures due to the use of an MRI probabilistic atlas. An SVM classifier is trained based on this set of features to identify the brain structures. We validated the approach using a gold standard real brain MRI data set. Comparison of the results with existing algorithms displays the promise of our approach. Funding:
|
Development of a CAD (Computer Assisted Detection) System to Detect Lung Nodules in CT Scans
|
Publication: PDF Authors: Eleonora Tommasi and Giorgio De Nunzio Institution: Departments of Physics and Material Science, University of Lecce, Italy Background/Purpose: We implemented a piece of software to segment lung tissue in CT thoracic images. We gave particular attention to the study of the ilhus pulmonis (region of entrance of bronchi and vessels in the lung parenchima). It is a delicate region in segmentation. We used 3D Slicer to visualize the different steps of the algorithm, checking their correctness. Through a segmentation technique, known as “region growing”, we obtained the delineation of the respiratory apparatus domain in the CT scans. This algorithm is completely three-dimensional. Its output consists in the 3D mask of the respiratory apparatus. We used 3D Slicer to visualize it. Then through an alternative version of region growing, based on a wavefront simulation algorithm, we obtained the segmentation of the external bronchi. We also used the opacity proprieties to put in evidence the internal structures of lungs (such as vessels). |
Dynamic Simulation of Joints Using Multi-Scale Modeling
|
Author: Trent M. Guess Institution: Mechanical Engineering, University of Missouri-Kansas City Background/Purpose:
Funding:
|
Quasi-isometric Flattening of Curved Surfaces for Medical Imaging
|
Researchers: Yehoshua Y. Zeevi, Emil Saucan and Eli Appleboim Institution: Department of Electrical-Engineering, Technion-Israel Institute of Technology, Haifa, Israel. Description: CT/MRI-scans of human brain-cortex/colon are flattened in a quasi-isometric manner in order to keep angles/length/area as minimally distorted as possible. 3D-surfaces are created out of real data scans, using 3D Slicer . Funding:
|
A Translation Station for Imaging
|
Publication: RSNA 2006; Authors: P Mongkolwat, T Lechner, A Kogan, S Talbot, B Johnson, D S Channin Institution: Imaging Informatics Section, Northwestern University, Chicago, IL 60611, USA Background/Purpose: Abstract: while providing clinical feedback to the basic sciences. In imaging informatics, this means enabling collaborations between radiologists and computer scientists. Commercial workstations do not allow for experimentation nor do they evolve fast enough. Research imaging packages, while innovative, do not provide for clinical activities or regulatory compliance. We added IHE workflow functionality, hanging protocols and other clinical functions to the 3D Slicer research platform. We leveraged the functionality and methodology of the Insight and Visualization Toolkits, and the DCM4CHE tools. A research mode allows for the evaluation of experimental algorithms against a PACS while protecting patient confidentiality. This combined functionality allows researchers to develop algorithms against clinical cases while at the same allowing clinicians to explore and validate these developments. |
User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability.
|
Publication: Neuroimage. 2006 Jul 1;31(3):1116-28. PMID: 16545965 | PDF Authors: Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. Institution: Penn Image Computing and Science Laboratory, Department of Radiology, University of Pennsylvania, PA 19104-6274, USA. Background/Purpose: Active contour segmentation and its robust implementation using level set methods are well-established theoretical approaches that have been studied thoroughly in the image analysis literature. Despite the existence of these powerful segmentation methods, the needs of clinical research continue to be fulfilled, to a large extent, using slice-by-slice manual tracing. To bridge the gap between methodological advances and clinical routine, we developed an open source application called ITK-SNAP, which is intended to make level set segmentation easily accessible to a wide range of users, including those with little or no mathematical expertise. This paper describes the methods and software engineering philosophy behind this new tool and provides the results of validation experiments performed in the context of an ongoing child autism neuroimaging study. The validation establishes SNAP intrarater and interrater reliability and overlap error statistics for the caudate nucleus and finds that SNAP is a highly reliable and efficient alternative to manual tracing. Analogous results for lateral ventricle segmentation are provided. Funding:
|
 Two and three-dimensional views of the caudate nucleus. Coronal slice of the caudate: original T1-weighted MRI (left) and overlay of segmented structures (middle). Right and left caudate are shown shaded in green and red; left and right putamen are sketched in yellow, laterally exterior to the caudates. The nucleus accumbens is sketched in red outline. Note the lack of contrast at the boundary between the caudate and the nucleus accumbens, and the fine-scale cell bridges between the caudate and the putamen. At right is a 3D view of the caudate and putamen relative to the lateral ventricles. |
Habitual Use of the Primate Forelimb is Reflected in the Material Properties of Subchondral Bone in the Distal Radius
|
Publication: J Anat. 2006 Jun; 208(6):659-670. PMID: 16761969 | PDF Authors: Carlson KJ, Patel BA. Institution: Department of Anatomical Sciences, School of Medicine, Stony Brook University, USA. Background/Purpose: Bone mineral density is directly proportional to compressive strength, which affords an opportunity to estimate in vivo joint load history from the subchondral cortical plate of articular surfaces in isolated skeletal elements. Subchondral bone experiencing greater compressive loads should be of relatively greater density than subchondral bone experiencing less compressive loading. Distribution of the densest areas, either concentrated or diffuse, also may be influenced by the extent of habitual compressive loading. We evaluated subchondral bone in the distal radius of several primates whose locomotion could be characterized in one of three general ways (quadrupedal, suspensory or bipedal), each exemplifying a different manner of habitual forelimb loading (i.e. compression, tension or non-weight-bearing, respectively). We employed computed tomography osteoabsorptiometry (CT-OAM) to acquire optical densities from whichfalse-colour maps were constructed. The false-colour maps were used to evaluate patterns in subchondral density (i.e. apparent density). Suspensory apes and bipedal humans had both smaller percentage areas and less well-defined concentrations of regions of high apparent density relative to quadrupedal primates. Quadrupedal primates exhibited a positive allometric effect of articular surface size on high-density area, whereas suspensory primates exhibited an isometric effect and bipedal humans exhibited no significant relationship between the two. A significant difference between groups characterized by predominantly compressive forelimb loading regimes vs. tensile or non-weight-bearing regimes indicates that subchondral apparent density in the distal radial articular surface distinguishes modes of habitually supporting of body mass. Funding:
|
Appearance of the Levator Ani Muscle Subdivisions in Magnetic Resonance Images
|
Publication: Obstet Gynecol. 2006 May;107(5):1064-9. PMID: 16648412 | PDF Authors: Rebecca U. Margulies, Md, Yvonne Hsu, Md, Rohna Kearney, Mrcog, Tamara Stein, Phd, Wolfgang H. Umek, Md, and John O. L. DeLancey, Md Institution: University of Michigan, Ann Arbor, Michigan; University College Hospital, London, United Kingdom; and Medical University Vienna, Vienna, Austria. Background/Purpose: Identify and describe the separate appearance of 5 levator ani muscle subdivisions seen in axial, coronal, and sagittal magnetic resonance imaging (MRI) scan planes. Magnetic resonance scans of 80 nulliparous women with normal pelvic support were evaluated. Characteristic features of each Terminologia Anatomica–listed levator ani component were determined for each scan plane. Muscle component visibility was based on pre- established criteria in axial, coronal, and sagittal scan planes: 1) clear and consistently visible separation or 2) different origin or insertion. Visibility of each of the levator ani subdivisions in each scan plane was assessed in 25 nulliparous women. In the axial plane, the puborectal muscle can be seen lateral to the pubovisceral muscle and decussating dorsal to the rectum. The course of the puboperineal muscle near the perineal body is visualized in the axial plane. The coronal view is perpendicular to the fiber direction of the puborectal and pubovisceral muscles and shows them as “clusters” of muscle on either side of the vagina. The sagittal plane consistently demonstrates the puborectal muscle passing dorsal to the rectum to form a sling that can consistently be seen as a “bump.” This plane is also parallel to the pubovisceral muscle fiber direction and shows the puboperineal muscle. The subdivisions of the levator ani muscle are visible in MRI scans, each with distinct morphology and characteristic features. Funding:
|
Virtual Cystoscopy - A Surgical Planning and Guidance Tool
|
Publication: Arch Ital Urol Androl. 2006 Mar;78(1):23-4. PMID: 16752884 Authors: Braticevici B, Onu M, Bengus F. Institution: Clinical Hospital, Bucharest, Romania. Background/Purpose: Image guided-surgery systems facilitates surgical planning phases of endoscopic procedures. In this paper, we used a software package for 3D surface model generation and vizualization of the urinary bladder, based on magnetic resonance (MR) cross sectional images of the pelvis. The patients group consisted in 6 patients diagnosed with urinary bladder tumour. They were submitted to MRI exam. Twelve consecutive cross sectional images of the pelvis were aquired (TR (repetition time) = 600 msec, TE (echo time) = 19 msec, slice thickness = 6 divided by 7 mm, FOV (field of view) = 36 cm. All these images were transferred to a personal computer running the 3D Slicer software. We obtained, for each patient, a 3D model of the pelvis including the urinary bladder. In This way, the surgical enviroment was simulated and we are able to investigate the bladder by virtual cystoscopy. The virtual endoscopy may be used as a tool in the preoperative training and in surgical planning. |
Measurement of the Pubic Portion of the Levator ani Muscle in Women with Unilateral Defects in 3D Models from MR Images
|
Publication: Int J Gynaecol Obstet. 2006 Mar;92(3):234-41. PMID: 16442111 | PDF Authors: Chen L, Hsu Y, Ashton-Miller JA, DeLancey JO Institution: Department of Biomedical Engineering, University of Michigan, Ann Arbor, Michigan 48109-2125, USA. Background/Purpose: Develop a method to quantify the cross-sectional area of the pubic portion of the levator ani muscle, validate the method in women with unilateral muscle defects, and report preliminary findings in those women. Multi-planar proton density magnetic resonance images of 12 women with a unilateral defect in the pubic portion of their levator ani were selected from a larger study of levator ani muscle anatomy in women with and without genital prolapse. Three-dimensional bilateral models of the levator ani were reconstructed (using 3D Slicer, version 2.1b1) and divided into iliococcygeal and pubic portions. Muscle cross-sectional areas were calculated at four equally spaced locations perpendicular to a line drawn from the pubic origin to the visceral insertion using the I-DEAS computer modeling software. The cross-sectional area of the muscle on the side with the defect was smaller than the normal side at all the four locations. The average bilateral difference was up to 81% at location 1 (nearest pubic origin). Almost all of the volume difference (13.7%, P=0.0004) was attributable to a reduction in the pubic portion (24.6%, P<0.0001), not the iliococcygeal portion (P=0.64), of the muscle. A method was developed to quantify cross-sectional area of the pubic portion of the levator ani perpendicular to the intact muscle direction. Significant bilateral cross-sectional area differences were found between intact and defective muscles in women with a unilateral defect. Funding:
|
Finite-element-method (FEM) Model Generation of Time-resolved 3D Echocardiographic Geometry Data for Mitral-valve Volumetry
|
Publication: Biomed Eng Online. 2006 Mar 3;5:17. PMID: 16512925 | PDF Authors: Verhey JF, Nathan NS, Rienhoff O, Kikinis R, Rakebrandt F, D'Ambra MN. Institution: MVIP ImagingProducts GmbH, Nörten-Hardenberg, Germany. Background/Purpose: Mitral Valve (MV) 3D structural data can be easily obtained using standard transesophageal echocardiography (TEE) devices but quantitative pre- and intraoperative volume analysis of the MV is presently not feasible in the cardiac operation room (OR). Finite element method (FEM) modelling is necessary to carry out precise and individual volume analysis and in the future will form the basis for simulation of cardiac interventions. METHOD: With the present retrospective pilot study we describe a method to transfer MV geometric data to 3D Slicer 2 software, an open-source medical visualization and analysis software package. A newly developed software program (ROIExtract) allowed selection of a region-of-interest (ROI) from the TEE data and data transformation for use in 3D Slicer. FEM models for quantitative volumetric studies were generated. ROI selection permitted the visualization and calculations required to create a sequence of volume rendered models of the MV allowing time-based visualization of regional deformation. Quantitation of tissue volume, especially important in myxomatous degeneration can be carried out. Rendered volumes are shown in 3D as well as in time-resolved 4D animations. The visualization of the segmented MV may significantly enhance clinical interpretation. This method provides an infrastructure for the study of image guided assessment of clinical findings and surgical planning. For complete pre- and intraoperative 3D MV FEM analysis, three input elements are necessary: 1. time-gated, reality-based structural information, 2. continuous MV pressure and 3. instantaneous tissue elastance. The present process makes the first of these elements available. Volume defect analysis is essential to fully understand functional and geometrical dysfunction of but not limited to the valve. 3D Slicer was used for semi-automatic valve border detection and volume-rendering of clinical 3D echocardiographic data. FEM based models were also calculated. A Philips/HP Sonos 5500 ultrasound device stores volume data as time-resolved 4D volume data sets. Data sets for three subjects were used. Since 3D Slicer does not process time-resolved data sets, we employed a standard movie maker to animate the individual time-based models and visualizations. Calculation time and model size were minimized. Pressures were also easily available. We speculate that calculation of instantaneous elastance may be possible using instantaneous pressure values and tissue deformation data derived from the animated FEM. Funding:
|
 Ultrasound data of after preprocessing. The ultrasound data are shown from patient 1. Shown is a standard visualization of one time step during the heart cycle with 3D Slicer software. The three planes for each spatial direction are shown in the lower three frames. In the frame above one plane is shown together with the set of four fiducials. These four points are defined for all time steps. |
Range of Curvilinear Distraction Devices Required for Treatment of Mandibular Deformities
|
Publication: J Oral Maxillofac Surg. 2006 Feb;64(2):259-64. PMID: 16413898 | PDF Authors: Ritter L, Yeshwant K, Seldin EB, Kaban LB, Gateno J, Keeve E, Kikinis R, Troulis MJ Institution: Department of Oral and Maxillofacial Surgery, Massachusetts General Hospital, Boston, MA, USA. Background/Purpose: The purpose of this study was to determine the range of fixed trajectory curvilinear distraction devices required to correct a variety of severe mandibular deformities. Preoperative computed tomography (CT) scans from 18 patients with mandibular deformities were imported into a CT-based software program (Osteoplan). Three-dimensional virtual models of the individual skulls were made with landmarks to track movements. An ideal treatment plan was created for each patient. Upper and lower boundaries for the dimensions of curvilinear distractors were established based on manufacturing and geometric constraints. Then, anatomically acceptable distractor attachment points were identified on the models using proximal and distal grids. Treatment plans were simulated for a series of distractors with varying radii of curvature, elongations (arc-length of device), and placements along the grids. The outcomes using these distractors were compared with the ideal treatment plans. Discrepancies were quantified in millimeters by comparing landmarks in the simulated versus ideal movements. Approximately 400,000 simulated 3-dimensional movements, based on the distractor parameters and variations in placement were computationally evaluated for the 18 cases. It was determined that, by varying distractor placement, a family of 5 distractors, with 3, 5, 7, and 10 cm radii of curvature and a straight-line device, could be used to treat all 18 cases to within 1.8 mm of error. The results of this study indicate that a family of 5 curvilinear distractors may suffice to treat a broad range of mandibular deformities. |
Preliminary Study on Digitized Nasal and Temporal Bone Anatomy
|
Publication: Clin Anat. 2006 Jan;19(1):32-6. PMID: 16283638 | PDF Authors: Li XP, Han DM, Xia Y, Zhou GH Institution: Beijing Tongren Hospital Affiliated with Capital University of Medical Sciences, ENT Department, People's Republic of China. Background/Purpose: The purpose of this study was to explore a feasible method for the reconstruction of the nasal and temporal bone structures of the Chinese virtual human project and provide a more accurate and facilitated way view them three-dimensionally (3D). The 3D Slicer software was used to reconstruct the anatomic structures of the human nose and temporal bone. Segmentation and extraction of the contours of the ROI (region of interest) in each single slice were conducted and the processed volume data was transferred into the 3D Slicer. After resegmentation, a set of labeled maps of the ROI were produced. Based on these maps, the 3D surface models of the tissues of interest were constructed. Four groups of paranasal sinuses, nasal septa, middle and inferior turbinates, temporal bones, tympanic cavities, mastoid air cells, sigmoid sinuses, and internal carotid arteries were reconstructed successfully. These models show spatial relationships and orientation between them. The results show that the 3D Slicer may be used for the 3D visualization of parts of anatomic structures in the nose and temporal bone based on the first Chinese virtual human data, and thus, can facilitate the observation and understanding of the anatomic structures in this area. Funding:
|
Developmental Response to Cold Stress in Cranial Morphology of Rattus: Implications for the Interpretation of Climatic Adaptation in Fossil Hominins
|
Publication: Proceedings of the Royal Society B: Biological Sciences 2006; 273:2605-2610. | PDF Authors: Todd C. Rae, Una Strand Viðarsdottir, Nathan Jeffery A. Theodore Steegmann Jr Institution: Evolutionary Anthropology Research Group, Department of Anthropology, University of Durham, 43 Old Elvet, Durham DH1 3HN, UK Background/Purpose: Adaptation to climate occupies a central position in biological anthropology. The demonstrable relationship between temperature and morphology in extant primates (including humans) forms the basis of the interpretation of the Pleistocene hominin Homo neanderthalensis as a cold-adapted species. There are contradictory signals, however, in the pattern of primate craniofacial changes associated with climatic conditions. To determine the direction and extent of craniofacial change associated with temperature, and to understand the proximate mechanisms underlying cold adaptations in vertebrates in general, dry crania rom previous experiments on cold- and warm-reared rats were investigated using computed tomography scanning and three-dimensional digitization of cranial landmarks. Aspects of internal and external cranial morphology were compared using standard statistical and geometric morphometric techniques. The results suggest that the developmental response to cold stress produces subtle but significant changes in facial shape, and a relative decrease in the volume of the maxillary sinuses (and nasal cavity), both of which are independent of the size of the skull or postcranium. These changes are consistent with comparative studies of temperate climate primates, but contradict previous interpretations of cranial morphology of Pleistocene Hominini. Funding:
|
A ceratopsid dinosaur parietal from New Mexico and its implications for ceratopsid biogeography and systematics
|
Publication: Journal of Vertebrate Paleontology 2006; 26:1018-1020. | PDF Authors: A. A. Farke, T. E. Williamson Institution: Department of Anatomical Sciences, Stony Brook University, N.Y. Background/Purpose: Ceratopsid (horned dinosaur) cranial material is relatively common in the Upper Cretaceous Naashoibito Member of the Kirtland Formation, San Juan Basin, New Mexico. However, the specimens are fragmentary, making identification problematic. Previously, it has been suggested that the Naashoibito specimens represent Torosaurus utahensis, a chasmosaurine taxon originally known from the North Horn Formation of Utah. This identification has been used to support a Lancian age for the Naashoibito Member and the Alamo Wash local fauna of the Kirtland Formation (e.g., Lehman, 1981; Lucas et al., 1987). Others (e.g., Lehman, 1990) have synonomized T. utahensis with Torosaurus latus, a taxon otherwise known only from the late Maastrichtian Lance and Hell Creek formations. Recent work has upheld the validity of T. utahensis (Sullivan et al., 2005). However, Sullivan and colleagues (2005) have also argued that no previously described ceratopsid specimens from the Naashoibito Member are identifiable beyond the level of Chasmosaurinae indeterminate and therefore are not useful for assessing biostratigraphic correlations and ceratopsid biogeography. Here we describe a partial chasmosaurine ceratopsid parietal from the Naashoibito Member of the Kirtland Formation. Although other fragmentary parietals have been recovered from this unit (e.g., Lehman, 1981; Lucas et al., 1987), the new specimen exhibits unusual morphology: an epoccipital element positioned along the midline. This feature has previously been reported only in Triceratops, a taxon not known from south of the Denver Basin. The occurrence of such a feature in a ceratopsid from New Mexico has implications for ceratopsian systematics and biogeography. Funding:
|
2005
Magnetic Resonance Imaging and 3-Dimensional Analysis of External Anal Sphincter Anatomy
|
Publication: Obstet Gynecol. 2005 Dec;106(6):1259-65. PMID: 16319250 | PDF Authors: Hsu Y, Fenner DE, Weadock WJ, DeLancey JO Institution: Pelvic Floor Research Group, Division of Gynecology, Department of Obstetrics and Gynecology, University of Michigan, Ann Arbor, Michigan 48109-0276, USA. Background/Purpose: To use magnetic resonance images of living women and 3-dimensional modeling software to identify the component parts and characteristic features of the external anal sphincter (EAS) that have visible separation or varying origins and insertions. METHODS: Detailed structural analysis of anal sphincter anatomy was performed on 3 pelvic magnetic resonance imaging (MRI) data sets selected for image clarity from ongoing studies involving nulliparous women. The relationships of anal sphincter structures seen in axial, sagittal, and coronal planes were examined using the 3D Slicer 2.1b1 software program. The following were requirements for sphincter elements to be considered separate: 1) a clear and consistently visible separation or 2) a different origin or insertion. The characteristic features identified in this way were then evaluated in images from an additional 50 nulliparas for the frequency of feature visibility. There were 3 components of the EAS that met criteria as being "separate" structures. The main body (EAS-M) is separated from the subcutaneous external anal sphincter (SQ-EAS) by a clear division that could be observed in all (100%) of the MRI scans reviewed. The wing-shaped end (EAS-W) has fibers that do not cross the midline ventrally, but have lateral origins near the ischiopubic ramus. This EAS-W component was visible in 76% of the nulliparas reviewed. Three distinct external anal sphincter components can be identified by MRI in the majority of nulliparous women. Funding:
|
 A. Inferior, left three-quarter view of model showing relationship of external anal sphincter complex to the bones and pelvic organs. B. Inferior, left three-quarter view of the external anal sphincter complex. External anal sphincter is in red; note the lateral wing portion of the external anal sphincter. Subcutaneous external anal sphincter is in orange. C. Posterior view of the external anal sphincter complex showing the circumferential nature of both the external anal sphincter and the subcutaneous external anal sphincter. U, urethra; B, bladder; V, vagina; LA, levator ani muscle; R, Rectum; EAS-M, main body of external anal sphincter; EAS-W, lateral wing portion of the external anal sphincter; SQ-EAS, subcutaneous external anal sphincter. |
Group-Slicer: a collaborative extension of 3D Slicer
|
Publication: J Biomed Inform. 2005 Dec;38(6):431-42. PMID: 16337568 | PDF Authors: Simmross-Wattenberg F, Carranza-Herrezuelo N, Palacios-Camarero C, Casaseca-de-la-Higuera P, Martin-Fernandez MA, Aja-Fernandez S, Ruiz-Alzola J, Westin CF, Alberola-Lopez C Institution: Laboratorio de Procesado de Imagen, ETSI Telecomunicacion, Universidad de Valladolid, 47011 Valladolid, Spain Background/Purpose: In this paper, we describe a first step towards a collaborative extension of the well-known 3D Slicer; this platform is nowadays used as a standalone tool for both surgical planning and medical intervention. We show how this tool can be easily modified to make it collaborative so that it may constitute an integrated environment for expertise exchange as well as a useful tool for academic purposes. Funding:
|
2D Rigid Registration of MR Scans using the 1D Binary Projections
|
Publication: Enformatika Transactions on Engineering, Computing and Technology, November 2005; 9:157-161. | PDF Author: Panos D. Kotsas Institution: Department of Automated Control and System Engineering, University of Sheffield, UK. Background/Purpose: This research deals with the application of a signal intensity independent registration criterion for 2D rigid body registration of medical images using 1D binary projections. The criterion is defined as the weighted ratio of two projections. The ratio is computed on a pixel per pixel basis and weighting is performed by setting the ratios between one and zero pixels to a standard high value. The mean squared value of the weighted ratio is computed over the union of the one areas of the two projections and it is minimized using the Chebyshev polynomial approximation using n=5 points. The sum of x and y parallel projections is used for translational adjustment and a range of parallel projections between 40-50 deg (depending on the orientation of the image) for rotational adjustment. MR-MR registration experiments were performed and gave mean errors well below 1deg and 1 pixel (0.12 deg and 0.47 pixels for the example given in Enformatika Publication). The method is to be extended for surface matching. |
Three-dimensional reconstruction and volumetry of intracranial haemorrhage and its mass effect
|
Publication: Neuroradiology. 2005 Jun;47(6):417-24. PMID: 15856213 | PDF Authors: Strik HM, Borchert H, Fels C, Knauth M, Rienhoff O, Bähr M, Verhey JF. Institution: Department of Neurology, Medical School, University of Göttingen, Germany. Background/Purpose: Intracerebral haemorrhage still causes considerable disability and mortality. The studies on conservative and operative management are inconclusive, probably due to inexact volumetry of the haemorrhage. We investigated whether three-dimensional (3D), voxel-based volumetry of the haemorrhage and its mass effect is feasible with routine computed tomography (CT) scans. The volumes of the haemorrhage, ventricles, midline shift, the intracranial volume and ventricular compression in CT scans of 12 patients with basal ganglia haemorrhage were determined with the 3D Slicer software. Indices of haemorrhage and intracranial or ventricular volume were calculated and correlated with the clinical data. The intended measures could be determined with an acceptable intra-individual variability. The 3D volumetric data tended to correlate better with the clinical course than the conventionally assessed distance of midline shift and volume of haemorrhage. 3D volumetry of intracranial haemorrhage and its mass effect is feasible with routine CT examination. Prospective studies should assess its value for clinical studies on intracranial space-occupying diseases. |
Vaginal Thickness, Cross-Sectional Area, and Perimeter in Women with and Those without Prolapse
|
Publication: Obstet Gynecol. 2005 May;105(5 Pt 1):1012-7. PMID: 15863538 | PDF Authors: Hsu Y, Chen L, Delancey JO, Ashton-Miller JA Institution: Division of Gynecology, Department of Obstetrics and Gynecology, University of Michigan Medical School, Ann Arbor, Michigan, USA. Background/Purpose: Use axial magnetic resonance imaging to test the null hypothesis that no difference exists in apparent vaginal thickness between women with and those without prolapse. Magnetic resonance imaging studies of 24 patients with prolapse at least 2 cm beyond the introitus were selected from an ongoing study comparing women with prolapse with normal control subjects. The magnetic resonance scans of 24 women with prolapse (cases) and 24 women without prolapse (controls) were selected from those of women of similar age, race, and parity. The magnetic resonance files were imported into an experimental modeling program, and 3-dimensional models of each vagina were created. The minimum transverse plane cross-sectional area, mid-sagittal plane diameter, and transverse plane perimeter of each vaginal model were calculated. Neither the mean age (cases 58.6 years ± standard deviation [SD] 14.4 versus controls 59.4 years ± SD 13.2) nor the mean body mass index (cases 24.1 kg/m2± SD 3.3, controls 25.7 kg/m2± SD 3.7) differed significantly between groups. Minimum mid-sagittal vaginal diameters did not differ between groups. Patients with prolapse had larger minimum vaginal cross-sectional areas than controls (5.71 cm2± standard error of the mean [SEM] 0.25 versus 4.76 cm2± SEM 0.20, respectively; P = .005). The perimeter of the vagina was also larger in the prolapse group (11.10 cm ± SEM 0.24) compared with controls (9.96 cm ± SEM 0.22) P = .001. Subgroup analysis of patients with endogenous or exogenous estrogen showed prolapse patients had larger vaginal cross-sectional area (P = .030); in patients without estrogen group differences were not significant (P = .099). Vaginal thickness is similar in women with and those without pelvic organ prolapse. The vaginal perimeter and cross-sectional areas are 11% and 20% larger in prolapse patients, respectively. Estrogen status did not affect differences found between groups. Funding:
|
 Axial slices at 5-mm intervals arranged caudal to cephalad starting from the image in the upper left (image 0). Vaginal tracings were made from above the level of the vestibular bulbs (VB), represented by asterisks (*), caudally (image 0) to below where the cervix (C) could be seen (image -4.0). U, urethra; V, vagina; R, rectum; B, bladder. |
Registration and Fusion of CT and MRI of the Temporal Bone
|
Publication: J Comput Assist Tomogr. 2005 May-Jun;29(3):305-10. PMID: 15891495 | PDF Authors: Bartling S, Peldschus K, Rodt T, Kral F, Matthies H, Kikinis R, Becker H. Institution: Department of Neuroradiology, Hannover Medical School, Hannover, Germany. Background/Purpose: CT and MRI of the skull base and the Temporal provides complimentary information. A registration and fusion of CT and MRI may improve diagnosis, because features only seen in one modality can be assessed with respect to the information gathered from the other modality. A manual, rigid registration method - basically overlaying two volumes in one space - as implemented in 3D Slicer was used. Threshold based fusion methods resulted in MRI overlaying co-registered CT. Landmarks have been used to assess target registration error - registration accuracy was within the range of the voxel size of either modality. A range of different pathological conditions was used to demonstrate the potential benefits of this methods. |
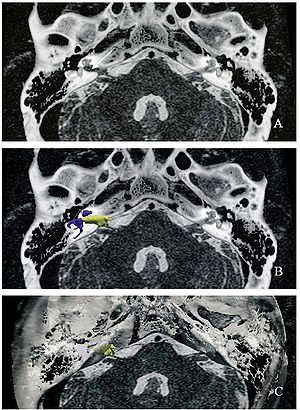 Acoustic schwannoma in the right internal auditory canal (arrow) in registered and fused CT and MRI of the temporal bone. For surgical planning, the MRI-given tumor (yellow) and critical CT-given surgical landmarks such as the labyrinth (blue) and the facial nerve canal (gray) as well as the bony skull base (white) can be displayed together. |
Open-configuration MR-guided Microwave Thermocoagulation Therapy for Metastatic Liver Tumors from Breast Cancer
|
Publication: Breast Cancer. 2005;12(1):26-31. PMID: 15657520 | PDF Authors: Abe H, Kurumi Y,Naka S, Shiomi H, Umeda T, Naitoh H, Endo Y, Hanasawa K, Morikawa S, Tani T. Institution: Division of General Surgery, Department of Surgery, Shiga University of Medical Science, Seta-Tsukinowa, Otsu, Shiga, 520-2192, Japan. Background/Purpose: Liver metastases from breast cancer are associated with a poor prognosis, however, local control with microwave thermocoagulation therapy has been used in certain subgroups of these patients in the past decade. In this study, open-configuration magnetic resonance (MR) -guided microwave thermocoagulation therapy was used for metastatic liver tumors from breast cancer, and the efficacy of this treatment was assessed. METHODS: Between June 2000 and April 2004, we used MR-guided microwave thermocoagulation therapy on 11 nodules in 8 patients with metastatic liver tumors from breast cancer. The procedure was carried out under general anesthesia. A 0.5 T open-configuration MR system and a microwave coagulator were used. Near-real-time MR images and real-time temperature images were collected and displayed on the monitor. The MR-compatible thoracoscope was used and combined with MR imaging guidance. Navigation software, a 3D Slicer, was installed and customized. The customized navigation software displayed near-real-time MR images. The percutaneous puncture into the tumors was successful in all cases. No mortality or major complications occurred as a result of the procedures. Five of the 8 patients are alive with new metastatic foci with a mean observation period of 25.9 months. We developed several devices to allow safe, easy, and accurate MR-guided microwave thermocoagulation therapy of liver tumors. Open-configuration MR-guided microwave thermocoagulation therapy appears to be a feasible method for tumor ablation of metastatic liver tumors from breast cancer. |
Quantification of Airway Diameters and 3D Airway Tree Rendering from Dynamic Hyperpolarized 3He Magnetic Resonance Imaging
|
Publication: Magn Reson Med. 2005 Feb;53(2):474-8. PMID: 15678546 | PDF Authors: Tina A. Lewis, Yang-Sheng Tzeng, Erin L. McKinstry, Angela C. Tooker, Kwansoo Hong, Yanping Sun, Joey Mansour, Zachary Handler, and Mitchell S. Albert Institution: Brigham and Womem's Hospital, Harvard Medical School, Boston, MA, USA. Background/Purpose: As another step toward extracting quantitative information from hyperpolarized 3He MRI, airway diameters in humans were measured from projection images and multislice images of the lungs. Values obtained were in good agreement with the Weibel lung morphometry model. The measurement of airway caliber can now be achieved without the use of ionizing radiation. Furthermore, it was demonstrated that 3D airway tree renderings could be constructed from the multislice data. Both the measurement of airway diameters and the rendering of 3D airway information hold promise for the clinical assessment of bronchoconstrictive diseases such as asthma and the associated evaluation of treatment effectiveness. Work is being done to address the uncertainties of the manually intensive methods we have developed. Funding:
|
A finite element method model to simulate laser interstitial thermo therapy in anatomical inhomogeneous regions
|
Publication: Biomed Eng Online. 2005 Jan 4;4(1):2. PMID: 15631630 | PDF Authors: Mohammed Y, Verhey JF. Institution: Department of Medical Informatics, University of Goettingen, Goettingen, Germany. Background/Purpose: Laser Interstitial ThermoTherapy (LITT) is a well established surgical method. The use of LITT is so far limited to homogeneous tissues, e.g. the liver. One of the reasons is the limited capability of existing treatment planning models to calculate accurately the damage zone. The treatment planning in inhomogeneous tissues, especially of regions near main vessels, poses still a challenge. In order to extend the application of LITT to a wider range of anatomical regions new simulation methods are needed. The model described with this article enables efficient simulation for predicting damaged tissue as a basis for a future laser-surgical planning system. Previously we described the dependency of the model on geometry. With the presented paper including two video files we focus on the methodological, physical and mathematical background of the model. |
 The geometry used. The left carotid artery is shown in this figure labelled with ca. The following letters indicate the orientation: s for superior, i for inferior, p for posterior, a for anterior, l for left, r for right. (a) is a photo of the human anatomy in the neck area. The carotid artery is shown here after moving the vein to the cranial direction. (b) shows the corresponding freehand 3D ultrasound dataset of the human neck region acquired axially. The 3D image in the top of (b) shows the 3D ultrasound volume together with the carotid artery segmented with 3D Slicer software [17]. The 3D model is displayed with 40% transparency. (c) displays the model used in the simulation approximated according to the geometry of the human neck shown in (a) and (b). The segmentation of the 3D ultrasound dataset in (b), is available as a video stream, too, showing the geometry of the carotid artery. |
The Application of DTI to Investigate White Matter Abnormalities in Schizophrenia
|
Publication: Ann. N.Y. Acad. Sci. 2005; 1064:134–148. PMID: 16394153 | PDF Authors: Marek Kubicki, Carl-Fredrik Westin, Robert W. McCarley, and Martha E. Shenton Institution: Clinical Neuroscience Division, Laboratory of Neuroscience, Boston VA Healthcare System–Brockton Division, Department of Psychiatry, Harvard Medical School, Brockton, Massachusetts, USA Background/Purpose: Schizophrenia is a serious and disabling mental disorder that affects approximately 1% of the general population, with often devastating effects on the psychological and financial resources of the patient, family, and larger community. The etiology of schizophrenia is not known, although it likely involves several interacting biological and environmental factors that predispose an individual to schizophrenia. However, although the underlying pathology remains unknown, it has been believed that brain abnormalities would ultimately be linked to the etiology of schizophrenia. This theory was rekindled in the 1970s, when the first computer-assisted tomography (CT) study showed enlarged lateral ventricles in schizophrenia. Since that time, there have been many improvements in MR acquisition and image processing, including the introduction of positron emission tomography (PET), followed by functional MR (fMRI), and diffusion tensor imaging (DTI). These advances have led to an appreciation of the critical role that brain abnormalities play in schizophrenia. While structural MRI has proven to be useful in investigating and detecting gray matter abnormalities in schizophrenia, the investigation of white matter has proven to be more challenging as white matter appears homogeneous on conventional MRI and the fibers connecting different brain regions cannot be appreciated. With the development of DTI, we are now able to investigate white matter abnormalities in schizophrenia. Funding:
|
 Fibers traveling through the splenium of the corpus callosum are shown, where fibers connecting the left and right occipital lobe are displayed in green, fibers interconnecting lateral temporal regions are displayed in red, and fibers connecting medial temporal regions are displayed in blue. Additionally, the genu of the corpus callosum is labeled as “G”, the body as “B”, the isthmus as “I”, and the splenium as “S”. |
2004
A Statistically Based Flow for Image Segmentation
|
Publication: Med Image Anal. 2004 Sep;8(3):267-74. PMID: 15450221 | PDF Authors: Pichon E, Tannenbaum A, Kikinis R. Institution: School of Electrical & Computer Engineering, Georgia Institute of Technology, Atlanta, GA, USA. Background/Purpose: In this paper we present a new algorithm for 3D medical image segmentation. The algorithm is versatile, fast, relatively simple to implement, and semi-automatic. It is based on minimizing a global energy defined from a learned non-parametric estimation of the statistics of the region to be segmented. Implementation details are discussed and source code is freely available as part of the 3D Slicer project. In addition, a new unified set of validation metrics is proposed. Results on artificial and real MRI images show that the algorithm performs well on large brain structures both in terms of accuracy and robustness to noise. Funding:
|
 Results on real and noisy simulated datasets (left and right respectively): (a) sagittal slice of real dataset and proposed segmentation (WM GM); (b) axial slice of artificial dataset and proposed segmentation (ventricle); (c) expert segmentation (gray) and proposed segmentation (white); (d) underlying ground truth (gray) and proposed segmentation (white); (e) rendered surface of proposed segmentation (WM GM); (f) rendered surface of proposed segmentation (ventricle). |
Decreases in Ventricular Volume Correlate with Decreases in Ventricular Pressure in Idiopathic Normal Pressure Hydrocephalus Patients who Experienced Clinical Improvement after Implantation with Adjustable Valve Shunts
|
Publication: Neurosurgery. 2004 Sep;55(3):582-92. PMID: 15335425 | PDF Authors: McConnell KA, Zou KH, Chabrerie AV, Bailey NO, Black PM. Institution: Department of Neurosurgery, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA. Background/Purpose: This retrospective study examined whether changes in ventricular volume correspond with changes in adjustable valve pressure settings in a cohort of patients who received shunts to treat idiopathic normal pressure hydrocephalus. We also examined whether these pressure-volume curves and other patient variables would co-occur with a positive clinical response to shunting. METHODS: We selected 51 patients diagnosed with idiopathic normal pressure hydrocephalus who had undergone implantation of a Codman Hakim programmable valve (Medos S.A., Le Locle, Switzerland). Clinical data were gathered from the patients. Funding:
|
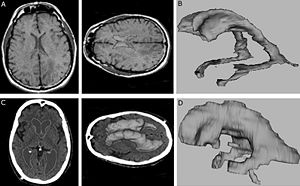 3D Slicer software segmentation of the ventricular system of a subject without hydrocephalus (A) used to produce the three-dimensional model of a normal, nonhydrocephalic ventricular system (B). C and D, 3D Slicer software segmentation of the ventricular system of a subject with INPH (C) used to produce the three-dimensional model of a hydrocephalic ventricular system (D). |
Spatial Motion Constraints in Medical Robot Using Virtual Fixtures Generated by Anatomy
|
Publication: Proceedings of the 2004 IEEE International Conference on Robotics & Automation, ICRA 2004.|PDF Authors: Ming Li, Russell H. Taylor Institution: Department of Computer Science, NSF Engineering Research Center for Computer Integrated Surgical Systems and Technology, Johns Hopkins University, Baltimore, MD, USA.
Funding:
|
Clinical validation of the normalized mutual information method for registration of CT and MR images in radiotherapy of brain tumors.
|
Publication: J Appl Clin Med Phys. 2004 Summer;5(3):66-79. PMID: 15753941 | PDF Authors: Veninga T, Huisman H, van der Maazen RW, Huizenga H. Institution: Department of Radiation Oncology, University Medical Center Nijmegen, The Netherlands. Background/Purpose: Image registration integrates information of different imaging modalities and has the potential to improve target volume determination in radiotherapy planning. This paper describes the implementation and validation of a 3D fully automated registration procedure in the process of radiotherapy treatment planning of brain tumors. 15 Patients with various brain tumors received CT and MR brain imaging before the start of radiotherapy. The normalized mutual information (NMI) method was used for image registration. Registration accuracy was estimated by performing statistical analysis of coordinate differences between CT and MR anatomical landmarks along the x-, y- and z-axes. Second, a visual validation protocol was developed to validate the quality of individual registration solutions and this protocol was tested in a series of 36 CT-MR registration procedures with intentionally applied registration errors. The mean coordinate differences between CT and MR landmarks along the x- and y-axes were in general within 0.5 mm. The mean coordinate differences along the z-axis were within 1.0 mm, which is of the same magnitude as the applied slice thickness in scanning. Second, the detection of intentionally applied registration errors by employment of a standardized visual validation protocol resulted in low false-negative and low false-positive rates. Application of the NMI method for the brain results in excellent automatic registration accuracy and the method has been incorporated in daily routine within our institute. A standardized validation protocol is proposed that ensures the quality of individual registrations by detecting registration errors with high sensitivity and specificity. This protocol is proposed for the validation of other linear registration methods. |
 For the purpose of individual quality assurance (QA) of image registration, an independent visual validation protocol was developed, using 3D Slicer, a freely available software program for visualization and processing of medical data. All image data were transferred to 3D Slicer in DICOM format. The validation protocol consists of a stepwise procedure for an estimate of translation errors along the three orthogonal axes (Trx, Try, and Trz) as well as rotation errors around these three axes (Rx, Ry, and Rz). |
Diffusion-tensor imaging–guided tracking of fibers of the pyramidal tract combined with intraoperative cortical stimulation mapping in patients with gliomas
|
Publication: J Neurosurg. 2004 Jul;101(1):66-72. PMID: 15255253 | PDF Authors: Berman JI, Berger MS, Mukherjee P, Henry RG. Institution: Department of Radiology, University of California at San Francisco, CA, USA.
Funding:
|
Abnormal Association between Reduced Magnetic Mismatch Field to Speech Sounds and Smaller Left Planum Temporale Volume in Schizophrenia
|
Publication: NeuroImage 2004; 22:720–727. PMID: 15193600 | PDF Authors: Hidenori Yamasue, Haruyasu Yamada, Masato Yumoto, Satoru Kamio, Noriko Kudo, Miki Uetsuki, Osamu Abe, Rin Fukuda, Shigeki Aoki, Kuni Ohtomo, Akira Iwanami Nobumasa Kato, Kiyoto Kasai Institution: Department of Neuropsychiatry, Graduate School of Medicine, University of Tokyo, Bunkyo, Tokyo 113-8655, Japan Background/Purpose: Schizophrenia is associated with language-related dysfunction. A previous study [Schizophr. Res. 59 (2003c) 159] has shown that this abnormality is present at the level of automatic discrimination of change in speech sounds, as revealed by magnetoencephalographic recording of auditory mismatch field in response to across-category change in vowels. Here, we investigated the neuroanatomical substrate for this physiological abnormality. Thirteen patients with schizophrenia and 19 matched control subjects were examined using magnetoencephalography (MEG) and high-resolution magnetic resonance imaging (MRI) to evaluate both mismatch field strengths in response to change between vowel /a/ and /o/, and gray matter volumes of Heschl's gyrus (HG) and planum temporale (PT). The magnetic global field power of mismatch response to change in phonemes showed a bilateral reduction in patients with schizophrenia. The gray matter volume of left planum temporale, but not right planum temporale or bilateral Heschl's gyrus, was significantly smaller in patients with schizophrenia compared with that in control subjects. Furthermore, the phonetic mismatch strength in the left hemisphere was significantly correlated with left planum temporale gray matter volume in patients with schizophrenia only. These results suggest that structural abnormalities of the planum temporale may underlie the functional abnormalities of fundamental language-related processing in schizophrenia. Funding:
|
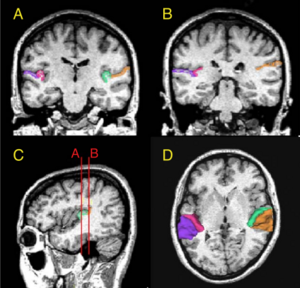 (A and B) Delineation of Heschl’s gyrus and planum temporale in a coronal slice in rostral and caudal part of regions of interest, respectively, based on MRI data of a control subject. The gray matter of Heschl’s gyrus is labeled green on subject left and wine-red on subject right. The gray matter of planum temporale is orange on subject left and violet on subject right. (C) Sagittal view of the Heschl’s gyrus and planum temporale in the left hemisphere. The coronal lines A and B correspond to the planes of panels A and B, respectively. (D) Three-dimensional reconstruction of Heschl’s gyrus and planum temporale gray matter superimposed on the axial plane. Each region is labeled using the same color as that in panels A, B and C. |
2003
Genetic contribution to cartilage volume in women: a classical twin study
|
Publication: Rheumatology (Oxford). 2003 Dec;42(12):1495-500. PMID: 12832711 | PDF Authors: David J. Hunter, Harold Snieder, Lyn March, and Philip N. Sambrook Institution: Institute of Bone and Joint Research, Royal North Shore Hospital, University of Sydney, Australia. Background/Purpose: A classical twin study was performed to assess the relative contribution of genetic and environmental factors to cartilage volume. METHODS: The subjects were 136 adult female twins: 31 monozygotic and 37 dizygotic twin pairs. The subjects had a T2-weighted fat-saturated sagittal gradient echo MRI performed of their right knee. Femoral, tibial and patella cartilage volumes were measured using 3D Slicer, a piece of software that facilitates semi-automatic segmentation, generation of three-dimensional surface models and quantitative analysis. The intraclass correlations were calculated, and maximum-likelihood model fitting was used to estimate genetic and environmental variance components. All variables were adjusted for age, BMI and femoral condyle size. The intraclass correlations for all of the cartilage volumes assessed were higher in monozygotic than dizygotic twin pairs. The heritabilities (95% confidence intervals) obtained from model fitting were: femoral, 61% (36-77%); tibial, 76% (56-87%); patella, 66% (47-79%); and total cartilage volume, 73% (51-85%). This study provides evidence for the importance of genetic factors in determining cartilage volume. Identifying heritability is the first step on the way to finding specific genes, which may improve our insight in the pathophysiology of cartilage disorders including the etiology of complex diseases such as osteoarthritis. Funding:
|
Advanced computer assistance for magnetic resonance-guided microwave thermocoagulation of liver tumors
|
Publication: Acad Radiol. 2003 Dec;10(12):1442-9. PMID: 14697012 | PDF Authors: Morikawa S, Inubushi T, Kurumi Y, Naka S, Sato K, Demura K, Tani T, Haque HA, Tokuda J, Hata N. Institution: Molecular Neuroscience Research Center, Shiga University of Medical Science, Seta Tsukinowa-cho, Ohtsu, Shiga 520-2192, Japan. Background/Purpose: The purpose of this study was to utilize computer assistance effectively for both easy and accurate magnetic resonance (MR) image-guided microwave thermocoagulation therapy of liver tumors. An open configuration MR scanner and a microwave coagulator at 2.45 GHz were used. Navigation software, a 3D Slicer, was customized to combine fluoroscopic MR images and preoperative MR images for the navigation. New functions to display MR temperature maps with simple parameter setting, and to record and display the coagulated areas by multiple microwave ablations in the 3-dimensional space (footprinting), were also introduced into the software. The VGA signal of the computer display was directly transferred to the surgeon's monitor. The customized software could be used for both accurate image navigation and convenient and easy temperature monitoring. Because repeated punctures and ablations are usually required in this procedure, the footprinting function made targeting of the tumors both easy and accurate and was quite effective in achieving the necessary and sufficient treatment. Furthermore, clear display on the surgeon's monitor, which was obtained by direct transfer of the VGA signal, enabled precise image navigation. The newly developed computer assistance was quite useful and helpful for this MR-guided procedure. |
The association of cartilage volume with knee pain
|
Publication: Osteoarthritis Cartilage. 2003 Oct;11(10):725-9. PMID: 13129691 | PDF Authors: Hunter DJ, March L, Sambrook PN. Institution: Institute of Bone and Joint Research, Royal North Shore Hospital, Sydney, Australia. Background/Purpose: Whilst the characteristic pathologic feature of OA is the loss of hyaline cartilage, prior studies have demonstrated a poor relationship between severity of reported knee pain and degree of radiographic change. The aim of this study was to examine the association between knee symptoms and MRI cartilage volume. DESIGN: A cross-sectional study was performed to assess the association between knee symptoms and MRI cartilage volume in an unselected, community based population. The subjects were 133 postmenopausal females. The subjects had a T2-weighted fat saturated sagittal gradient-echo MRI performed of their right knee. Femoral, tibial and patella cartilage volumes were measured using three-dimensional 3D Slicer, a software that facilitates semi-automatic segmentation, generation of 3D surface models and quantitative analysis. Qualitative data relating to symptoms, stiffness, pain, physical dysfunction and the quality of life using the WOMAC were recorded. The statistical analyses conducted to determine measures of association between knee pain/symptoms and cartilage volume were correlation, multiple regression and inter-quartile regression. Assessment of the association between patella cartilage volume and the WOMAC domains showed an inverse relationship between patella cartilage volume and pain, function and global score in a model including body mass index, physical activity and leg extensor power (all P=0.01). Inter-quartile regression comparing the lowest 25% with highest 25% patella cartilage volume demonstrated a stronger inverse relationship (P=0.005). This study suggests that alterations in patella volume are associated with pain, function and global scores of the WOMAC. In participants with more knee pain, there was an association with severity of patella cartilage reduction. Other MRI cartilage volume features were not strongly associated with WOMAC sub-scores. |
Three-dimensional reconstruction of magnetic resonance images of the anal sphincter and correlation between sphincter volume and pressure
|
Publication: Am J Obstet Gynecol. 2003 Jul;189(1):130-5. PMID: 12861151 | PDF Authors: Cornella JL, Hibner M, Fenner DE, Kriegshauser JS, Hentz J, Magrina JF. Institution: Mayo Graduate School of Medicine, USA. Background/Purpose: The purpose of this study was to assess the correlation between internal and external anal sphincter volumes and manometric anal pressures. Ten healthy nulliparous women underwent anal sphincter magnetic resonance imaging and anal manometry measurement. A 3-dimensional reconstruction of magnetic resonance images was accomplished with the use of 3D Slicer. Sphincter volumes were measured 3 times by the same observer for each of 10 patients. The intrarater reliability was measured with the use of the intraclass correlation coefficient (ICC = sigma2(patients)/(sigma 2(patients) + sigma2(error))) from a 2-way analysis of variance model with terms for patient and measurement trial. Measurements that were recorded on anal manometry included squeeze length, length of the high-pressure zone, and maximal resting and squeeze pressures. The mean volumes (± SD) were 18.77 ± 4.64 cm3, 13.82 ± 3.8 cm3, and 32.36 ± 8.37 cm3 for internal, external, and combined sphincters, respectively. Intrarater reliability was 98% for external sphincter volume (95% CI, 94%-99%), 98% for internal sphincter volume (95% CI, 94%-99%), and 99% for total volume (95% CI, 97%-100%). On the 3-dimensional images, the internal sphincter was found to be cylindric in shape, with an ellipse as a base. It is elongated in the anterior and posterior direction and flattened on the sides. The external sphincter was found to be funnel-shaped, being narrower caudad and widening in the cephalad direction. Similar to the internal sphincter, the external sphincter is elongated in the anteroposterior diameter. Volumes of the internal, external, and combined sphincters did not correlate with the maximum pressures at rest and squeeze. Correlations higher than r = 0.5 were observed for all 3 sphincter volume measurements versus high pressure zone at squeeze. The highest correlation, r = 0.66, was for internal sphincter volume versus high pressure zone at squeeze Three-dimensional reconstruction of the rectal sphincter musculature can be performed easily with 3-dimensional software. Measurements of the sphincter volumes have excellent intrarater reliability. Sphincter volumes do not correlate with pressures at rest or squeeze, but the internal sphincter volume correlates with the length of the high pressure zone at squeeze. Contrary to current generalized concepts, it is possible that the internal sphincter may play some role in generating the squeeze pressure. More research is necessary in applying 3-dimensional magnetic resonance image reconstruction in patients with different parity and continence status. Reconstruction of magnetic resonance images of the rectal sphincter musculature may prove to be beneficial in planning the treatment of patients with fecal incontinence. |
|
Publication: Acta Neurochir Suppl. 2003;85:121-5. PMID: 12570147 | PDF Authors: Nabavi A, Gering DT, Kacher DF, Talos IF, Wells WM, Kikinis R, Black PM, Jolesz FA. Institution: Department of Neurosurgery, University Kiel, Kiel, Germany. Background/Purpose: The introduction of MRI into neurosurgery has opened multiple avenues, but also introduced new challenges. The open-configuration intraoperative MRI installed at the Brigham and Women's Hospital in 1996 has been used for more than 500 open craniotomies and beyond 100 biopsies. Furthermore the versatile applicability, employing the same principles, is evident by its frequent use in other areas of the body. However, while intraoperative scanning in the SignaSP yielded unprecedented imaging during neurosurgical procedures their usage for navigation proved bulky and unhandy. To be fully integrated into the procedure, acquisition and display of intraoperative data have to be dynamic and primarily driven by the surgeon performing the procedure. To use the benefits of computer-assisted navigation systems together with immediate availability of intraoperative imaging we developed a software package. This 3D Slicer has been used routinely for biopsies and open craniotomies. The system is stable and reliable. Pre- and intraoperative data can be visualized to plan and perform surgery, as well as to accommodate for intraoperative deformations, "brain shift", by providing online data acquisition. Funding:
|
The virtual craniofacial patient: 3D jaw modeling and animation
|
Publication: Stud Health Technol Inform. 2003;94:65-71. PMID: 15455866 | PDF Authors: Enciso R, Memon A, Fidaleo DA, Neumann U, Mah J. Institution: Craniofacial Virtual Reality Laboratory, University of Southern California, CA, USA. Background/Purpose:Recent developments in technology and software are providing better data and methods to facilitate research in biomedical modeling and simulation. In the area of segmentation, original methods involved the time-consuming task of manually tracing structures from slice to slice. This process is now possible with significantly less interaction from the user. Programs such as 3D Slicer [6], Mimics (Materialise N.V., Heverlee, Belgium) and Amira 2.3 (TGS) provide semi-automatic image-processing-based segmentation and modeling from CT images. These programs use the Generalized Marching Cubes algorithm to create a polygonal wireframe mesh of the segmented area and export a model in STL or other formats. Funding:
|
2002
3D Visualisation of the Middle Ear and Adjacent Structures Using Reconstructed Multi-slice CT Datasets, Correlating 3D Images and Virtual Endoscopy to the 2D Cross-sectional Images
|
Publication: Neuroradiology. 2002 Sep;44(9):783-90. PMID: 12221454 | PDF Authors: Rodt T, Ratiu P, Becker H, Bartling S, Kacher DF, Anderson M, Jolesz FA, Kikinis R. Institution: Department of Neuroradiology, Hanover Medical School, Hanover, Germany. Background/Purpose: The 3D imaging of the middle ear facilitates better understanding of the patient's anatomy. Cross-sectional slices, however, often allow a more accurate evaluation of anatomical structures, as some detail may be lost through post-processing. In order to demonstrate the advantages of combining both approaches, we performed computed tomography (CT) imaging in two normal and 15 different pathological cases, and the 3D models were correlated to the cross-sectional CT slices. Reconstructed CT datasets were acquired by multi-slice CT. Post-processing was performed using the in-house software 3D Slicer, applying thresholding and manual segmentation. 3D models of the individual anatomical structures were generated and displayed in different colours. The display of relevant anatomical and pathological structures was evaluated in the greyscale 2D slices, 3D images, and the 2D slices showing the segmented 2D anatomy in different colours for each structure. Correlating 2D slices to the 3D models and virtual endoscopy helps to combine the advantages of each method. As generating 3D models can be extremely time-consuming, this approach can be a clinically applicable way of gaining a 3D understanding of the patient's anatomy by using models as a reference. Furthermore, it can help radiologists and otolaryngologists evaluating the 2D slices by adding the correct 3D information that would otherwise have to be mentally integrated. The method can be applied to radiological diagnosis, surgical planning, and especially, to teaching. |
2001
Serial Intraoperative Magnetic Resonance Imaging of Brain Shift
|
Publication: Neurosurgery. 2001 Apr;48(4):787-97. PMID: 11322439 | PDF Authors: Nabavi A, Black PM, Gering DT, Westin CF, Mehta V, Pergolizzi RS Jr, Ferrant M, Warfield SK, Hata N, Schwartz RB, Wells WM 3rd, Kikinis R, Jolesz FA. Institution: Division of Neurosurgery, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA. Background/Purpose: A major shortcoming of image-guided navigational systems is the use of preoperatively acquired image data, which does not account for intraoperative changes in brain morphology. The occurrence of these surgically induced volumetric deformations ("brain shift") has been well established. Maximal measurements for surface and midline shifts have been reported. There has been no detailed analysis, however, of the changes that occur during surgery. The use of intraoperative magnetic resonance imaging provides a unique opportunity to obtain serial image data and characterize the time course of brain deformations during surgery. The vertically open intraoperative magnetic resonance imaging system (SignaSP, 0.5 T; GE Medical Systems, Milwaukee, WI) permits access to the surgical field and allows multiple intraoperative image updates without the need to move the patient. We developed volumetric display software, the 3D Slicer, that allows quantitative analysis of the degree and direction of brain shift. For 25 patients, four or more intraoperative volumetric image acquisitions were extensively evaluated. Serial acquisitions allow comprehensive sequential descriptions of the direction and magnitude of intraoperative deformations. Brain shift occurs at various surgical stages and in different regions. Surface shift occurs throughout surgery and is mainly attributable to gravity. Subsurface shift occurs during resection and involves collapse of the resection cavity and intraparenchymal changes that are difficult to model. Brain shift is a continuous dynamic process that evolves differently in distinct brain regions. Therefore, only serial imaging or continuous data acquisition can provide consistently accurate image guidance. Furthermore, only serial intraoperative magnetic resonance imaging provides an accurate basis for the computational analysis of brain deformations, which might lead to an understanding and eventual simulation of brain shift for intraoperative guidance. Funding:
|
 Color-coded models for the evaluation of successive surface deformations. The brain surfaces were extracted from the 3D volume SPGRs for all time points. The surface displacement from stage to stage was automatically calculated and color-coded. The color encodes depths from red (0 mm) to green (2.5 cm) to blue (5 cm). The color-coding was projected onto the model of the brain at that respective stage of the surgery. |
2000
Image-guided Therapy and Intraoperative MRI in Neurosurgery
|
Publication: Minim Invasive Ther Allied Technol. 2000;9(3-4):277-86. PMID: 20156025 Authors: Nabavi A, Mamisch CT, Gering DT, Kacher DF, Pergolizzi RS, Wells WM 3rd, Kikinis R, Black PM, Jolesz FA. Institution: Department of Radiology, Magnetic Resonance Therapy, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA. Background/Purpose: Computer-assisted 3D planning, navigation and the possibilities offered by intra-operative imaging updates have made a large impact on neurological surgery. Three-dimensional rendering of complex medical image information, as well as co-registration of multimodal sources has reached a highly sophisticated level. When introduced into surgical navigation however, this pre-operative data is unable to account for intra-operative changes, ('brain-shift'). To update structural information during surgery, an open-configured, intra-operative MRI (Signa SP, 0.5 T) was realised at our institution in 1995. The design, advantages, limitations and current applications of this system are discussed, with emphasis on the integration of imaging into procedures. We also introduce our integrated platform for intra-operative visualisation and navigation, the 3D Slicer. Funding:
|