Difference between revisions of "Special:Badtitle/NS100:Introduction"
| Line 20: | Line 20: | ||
'''Slicer and Image-Guided Therapy''' | '''Slicer and Image-Guided Therapy''' | ||
| − | [[ | + | [[Image:Fedorov-Supercomputing2006-fig3.png|thumb|right|300 px|Overlaying MRI Images To Guide Decision-making When Resecting Tumor. See [http://slicer.spl.harvard.edu/Special:PubDB_View?dspaceid=44 Chrisochoides et al ICHPC 2006] for more info.]] |
With IRB clinical protocols appropriately created and managed, Slicer can be and has been involved in clinical trials. For image-guided therapy interventions, Slicer is frequently used to construct and visualize collections of MRI data that are available pre- and intraoperatively to allow for the acquiring of spatial coordinates for instrument tracking. In fact, Slicer has already played such a pivotal role in image-guided therapy, it could be thought of as growing up alongside that field. | With IRB clinical protocols appropriately created and managed, Slicer can be and has been involved in clinical trials. For image-guided therapy interventions, Slicer is frequently used to construct and visualize collections of MRI data that are available pre- and intraoperatively to allow for the acquiring of spatial coordinates for instrument tracking. In fact, Slicer has already played such a pivotal role in image-guided therapy, it could be thought of as growing up alongside that field. | ||
Revision as of 20:21, 25 July 2007
Overview
Slicer, or 3D Slicer, is open source software used for biomedical image analysis. Ever evolving as a modular platform in Windows, Linux or Darwin environments, Slicer's basic functionality includes:
- interactive visualization of images,
- manual editing,
- fusion and co-registering of data,
- automatic segmentation,
- analysis of diffuse tensor imaging data, and
- visualization of tracking information for image-guided procedures.
Slicer started as a project between the Surgical Planning Laboratory at the Brigham and Women's Hospital and the MIT Artificial Intelligence Laboratory in 1998 but is now in wide use in hundreds of applications. For one of those applications, image-guided therapy, it has been integral, helping to advance that field.
Given its versatility, Slicer has already been used in conjunction with other research tools, such as the image-guided navigation software, theSIGN. Such advances will continue in part through the expertise and effort of connected communities; namelyNAMIC, NAC,BIRN and NCIGTcommunities. These entities also serve as funding support, along with the NCRR, NIBIB,NIH Roadmap, NCI,NSF and the DOD as well as others.
Slicer 3 is the latest Slicer version that keeps in mind the needs of the person without a computer science background, whether he or she is a user or developer. Additionally, it preserves the core functionality of previous versions of Slicer. Newer features include: MRML load/save, viewing and display, command line modules (filtering, segmentation, registration, model building...), basic label map editor, EMSegmenter, completed overall GUI, color nodes, fiducials, linear transforms, model display, and model clipping.
Slicer modules and source code are available to the public under a BSD-style, open source licensing agreement under which there are no reciprocity requirements. This status makes the development of Slicer an endeavor that crosses research communities. With commonly-understood software components as its basis (Tcl, Tk, and VTK), the Slicer platform can be layered onto relatively easily with newly created modules. A Slicer "Wiki" acts as a laboratory notebook, if you will, tracking progress on independent and collaborative projects by researchers around the world.
Slicer and Image-Guided Therapy
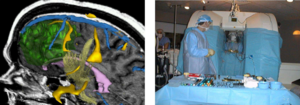
With IRB clinical protocols appropriately created and managed, Slicer can be and has been involved in clinical trials. For image-guided therapy interventions, Slicer is frequently used to construct and visualize collections of MRI data that are available pre- and intraoperatively to allow for the acquiring of spatial coordinates for instrument tracking. In fact, Slicer has already played such a pivotal role in image-guided therapy, it could be thought of as growing up alongside that field.
In addition to producing 3D models from conventional MRI images, Slicer has also been used to present information derived from fMRI (using MRI to assess blood flow in the brain related to neural or spinal cord activity), DTI (using MRI to measure the restricted diffusion of water in imaged tissue), and electrocardiography. For example, Slicer's DTI data analysis ability allows gradient images to convert to volumetric tensor data that can be visualized as glyphs or as trace representations of tract structures from interactively located seed coordinates from specified regions of interest.
For Slicer's application in neurosurgery, neuro-specific modules continue to be created. For example, the diffusion tensor imaging (DTI) release has the following features:
- DWI load/display,
- diffusion tensor estimation,
- tensor scalar calculation,
- tensor glyph display,
- interactive tractography, and
- the ability to generate tractography statistics.
By enabling DTI imaging, Slicer allows MRI to distinguish better between what's normal and abnormal as well as various parts of the brain in ways that conventional MRI images cannot. Such enabled functionality guides researchers to new understandings about the basis, diagnosis, and treatment of disease. To date the Slicer DTI modules have allowed researchers to compare the white matter of a normal brain to one with abnormalities associated with schizophrenia.