Modules:Volumes:Diffusion Editor-Documentation-3.6
Return to Slicer 3.6 Documentation
Module Name
Diffusion Editor
General Information
Module Type & Category
Type: Interactive
Category: Diffusion Imaging
Authors, Collaborators & Contact
- Author1: Kerstin Kessel
- Contact: kerstin@bwh.harvard.edu
Module Description
Overview of what the module does goes here.
Usage
Use Cases, Examples
This module is especially appropriate for these use cases:
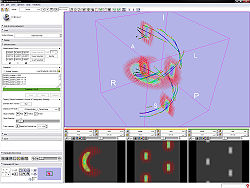
For DWI
- Use Case 1: Edit gradients manually or load existing gradients from file (.txt or .nhdr).
- Use Case 2: Edit the measurement frame manually or simply rotate/swap/invert columns by selecting them.
- Use Case 3: Test the parameters by estimating a tensor and displaying glyphs and tracts.
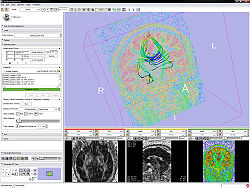
For DTI
- Use Case 2: Edit the measurement frame as described above.
- Use Case 3: Test by displaying glyphs and tracts.
Examples of the module in use:
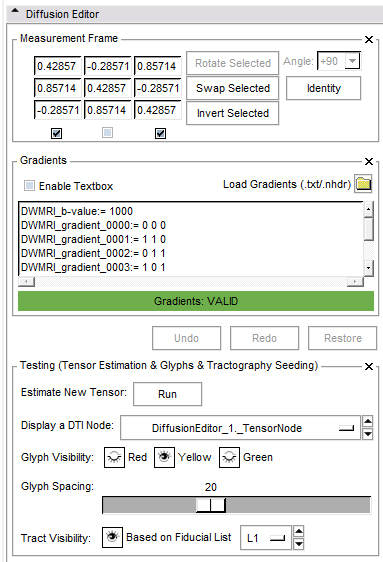
- Measurement frame:
- The determinant of the measurement frame has to be 1. This is checked by the editor.
- Invert: Select the columns you want to invert.
- Swap: Select two columns you want to swap.
- Rotate: Select one column you want to rotate by an angle you can choose from a given set of values or set yourself.
- Identity: Set the measurement frame to the identity matrix.
- Set your own values.
- Gradients frame:
- If the active volume is a DWI the editor will put the gradients in the text field; otherwise this frame is disabled.
- You can copy/paste your own gradients in the text field or change them manually.
- You can load gradients from a text file or .nhdr file. For the .txt file the format of gradients is easy, it can contain only values ( see example data ).
- Undo/Redo/Restore:
- Undo: Undo the last change of measurement/gradient values.
- Redo: Redo the last change of measurement/gradient values.
- Restore: All parameters are restored to original.
NOTE: You lose all previous changes when a new active volume is loaded or selected.
- Testing frame:
- Run tensor estimation. The new tensor node shows up in the DTI selector.
- Select a DTI node from the current mrml scene and for
- Glyph visibility: Select the planes on which you want to see glyphs and adjust the glyph spacing.
- Tract visibility: Add some fiducials and choose the fiducial list in the selector. Show tracts with Tractography Fiducial Seeding.
Tutorials
Links to tutorials explaining how to use this module:
- Tutorial 1
- Data Set 1
Quick Tour of Features and Use
A list panels in the interface, their features, what they mean, and how to use them. For instance:
|
Development
Notes from the Developer(s)
Algorithms used, library classes depended upon, use cases, etc.
Dependencies
Other modules or packages that are required for this module's use.
Tests
On the Dashboard, these tests verify that the module is working on various platforms:
- MyModuleTest1 MyModuleTest1.cxx
- MyModuleTest2 MyModuleTest2.cxx
Known bugs
Links to known bugs in the Slicer3 bug tracker
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Links to the module's source code:
Source code:
Doxygen documentation:
More Information
Acknowledgment
Include funding and other support here.
References
Publications related to this module go here. Links to pdfs would be useful.