Difference between revisions of "Modules:Volumes:Diffusion Editor-Documentation"
| Line 46: | Line 46: | ||
* Download example data: [[ media:DiffusionEditor_ExampleData.zip | DiffusionEditor_ExampleData.zip ]] | * Download example data: [[ media:DiffusionEditor_ExampleData.zip | DiffusionEditor_ExampleData.zip ]] | ||
| − | == Tutorials == | + | ===== Tutorials ===== |
<font color="Red"> TODO: Tutorials; add ExampleData </font> | <font color="Red"> TODO: Tutorials; add ExampleData </font> | ||
Revision as of 21:32, 1 May 2008
Home < Modules:Volumes:Diffusion Editor-DocumentationReturn to Slicer Documentation
Diffusion Editor
General Information
Module Type & Category
Type: Interactive
Category: Diffusion Imaging
Authors, Collaborators & Contact
- Author: Kerstin Kessel
- Supervisor: Steve Pieper, Ron Kikinis
Module Description
As the documentation of acquisition parameters in dicom data is not standardized, MRI scanners (Siemens, GE) handle gradients/bValues differently in their dicom headers. Because of that there is a big need to add/modify them manually.
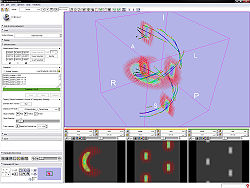
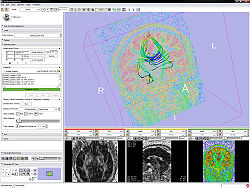
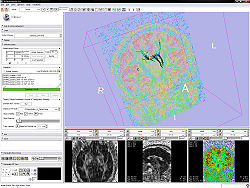
The Diffusion Editor is a 3D Slicer module that allows modifying parameters (gradients, bValues, measurement frame) of DWI data and provides a quick way to interpret them. For that it estimates a tensor and shows glyphs and tracts for visual exploration.
Current features of the Diffusion Editor
- For DWI
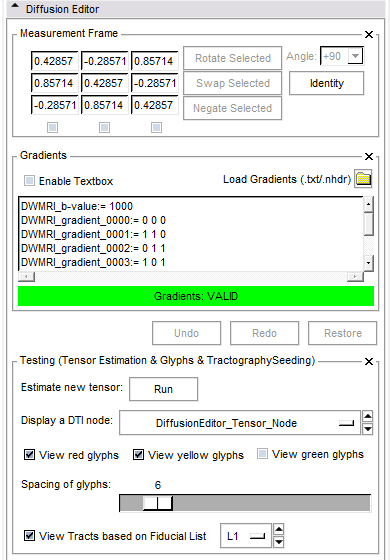
- Edit gradients manually or load existing gradients from file (.txt or .nhdr).
- Edit the measurement frame manually or simply rotate/swap/negate columns by selecting them.
- Test the parameters by estimating a tensor and displaying glyphs and tracts.
- For DTI
- Edit the measurement frame as described above.
- Test by displaying glyphs and tracts.
Usage
Examples & Tutorials
Preparation
- Download Slicer. The most recent stable Slicer version can be found here. To use the current version download the latest Slicer snapshot.
- You can also use SVN to checkout the trunk of the very latest version of Slicer. See here for more instructions on how to build Slicer.
- Download example data: DiffusionEditor_ExampleData.zip
Tutorials
TODO: Tutorials; add ExampleData
| title | used data | short description |
|---|---|---|
| Load | ___ | Load a DWI dicom data set in Slicer. |
| Measurement frame | ___ | Change the measurement frame by rotating, swapping, inverting or changing to identity. |
| Gradients | ___ | Change gradients manually or load them from a file. |
| Testing | ___ | Run tensor estimation and show glyphs and tracts;
Run second estimation with 90°rotation and switch between both tensors and see the impact of that change. |
Quick Tour of Features and Use
- Measurement frame:
- The determinant of the measurement frame has to be 1. This is checked by the editor.
- Invert: Select the columns you want to invert.
- Swap: Select two columns you want to swap.
- Rotate: Select one column you want to rotate by an angle you can choose from a given set of values or set yourself.
- Identity: Set the measurement frame to the identity matrix.
- Set your own values.
- Gradients frame:
- If the active volume is a DWI the editor will put the gradients in the text field; otherwise this frame is disabled.
- You can copy/paste your own gradients in the text field or change them manually.
- You can load gradients from a text file or .nhdr file. For the .txt file the format of gradients is easy, it can contain only values ( see example data ).
- Undo/Redo/Restore:
- Undo: Undo the last change of measurement/gradient values.
- Redo: Redo the last change of measurement/gradient values.
- Restore: All parameters are restored to original.
NOTE: You lose all previous changes when a new active volume is loaded or selected.
- Testing frame:
- Run tensor estimation. The new tensor node shows up in the DTI selector.
- Select a DTI node from the current mrml scene and for
- Glyph visibility: Select the planes on which you want to see glyphs and adjust the glyph spacing.
- Tract visibility: Add some fiducials and choose the fiducial list in the selector. Show tracts with Tractography Fiducial Seeding.
Development
Status / Implementation Progress
The editor is now successfully integrated in the Volumes module of the trunk version of Slicer3.
- measurement frame (gui/load/change/save): 100%
- gradients (gui/load/change/save): 100%
- undo/redo/restore mechanism: 100%
- testing frame: 100%
- tensor estimation with existing clm: 100%
- glyphs view in all three planes with the possibility to change spacing: 100%
- tractography fiducial seeding: 100%
- ctest: 50%
- documentation, tutorials (videos): 40%
- writing master thesis: 5% :-)
Known bugs
Follow this link to the Slicer3 bug tracker to see known bugs, or to report a new one. Please select the usability issue category when browsing or contributing:
http://na-mic.org/Mantis/main_page.php
Queries, feature requests and other feedback should be directed to slicer-devel |at| bwh.harvard.edu. You can also sign up for Slicers user and developer list to make sure you hear of new developments, releases and bug fixes.
Source code & documentation
Source code is available here:
http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerDiffusionEditorWidget.cxx http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerDiffusionEditorWidget.h
http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerDiffusionEditorLogic.cxx http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerDiffusionEditorLogic.h
http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerMeasurementFrameWidget.cxx http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerMeasurementFrameWidget.h
http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerGradientsWidget.cxx http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerGradientsWidget.h
http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerDWITestingWidget.cxx http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerDWITestingWidget.h
Links to documentation generated by doxygen:
http://www.na-mic.org/Slicer/Documentation/Slicer3/html/
More Information
Acknowledgement
This research project was part of a Master thesis (Diplomarbeit) in cooperation of the faculty of Medical Informatics, University of Heidelberg/Germany and the Surgical Planning Lab. It was supported by a grant of the Lions Club Heilbronn/Franken.
References
TODO: add link to thesis, papers...
Nrrd format: http://wiki.na-mic.org/Wiki/index.php/NAMIC_Wiki:DTI:Nrrd_format
DTMRI: http://wiki.na-mic.org/Wiki/index.php/Slicer3:DTMRI
DICOM for DWI and DTI : http://wiki.na-mic.org/Wiki/index.php/NAMIC_Wiki:DTI:DICOM_for_DWI_and_DTI