Difference between revisions of "Modules:VMTKLevelSetSegmentation"
| Line 72: | Line 72: | ||
===Known bugs=== | ===Known bugs=== | ||
| − | Follow this [http:// | + | Follow this [http://www.nitrc.org/tracker/?func=browse&group_id=196&atid=821 link] to the VMTK in 3D Slicer bug tracker. |
| − | |||
| − | |||
===Usability issues=== | ===Usability issues=== | ||
| Line 82: | Line 80: | ||
===Source code & documentation=== | ===Source code & documentation=== | ||
| − | + | The complete source code is available at a [http://www.nitrc.org/plugins/scmsvn/viewcvs.php/VmtkSlicerModule/?root=slicervmtklvlst NITRC SVN repository]. | |
| − | |||
| − | |||
| − | |||
== More Information == | == More Information == | ||
Revision as of 00:20, 16 July 2009
Home < Modules:VMTKLevelSetSegmentationReturn to Slicer 3.4 Documentation
Module Name
VMTKLevelSetSegmentation
General Information
Module Type & Category
Type: Interactive Scripted Module
Category: Segmentation, Extension
Authors, Collaborators & Contact
- Author: Daniel Haehn, University of Heidelberg
- Supervisor: Luca Antiga, Mario Negri Institute
- Contact: Daniel Haehn, haehn@bwh.harvard.edu
Module Description
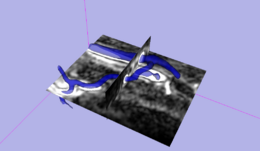
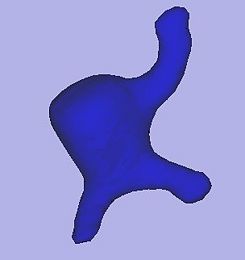
This module provides the level-set segmentation process of the Vascular Modeling Toolkit (http://www.vmtk.org) in 3D Slicer. The process targets manual segmentation of tubular and blob-like structures.
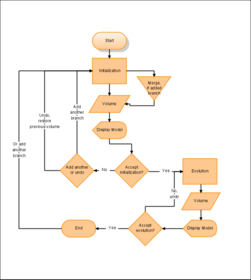
Segmentation using level sets consists of an initialization and an evolution step. The initialization step involves the description of a starting model within a region of interest. In the evolution step this initial deformable model then gets inflated to match the contours of the targeted volume.
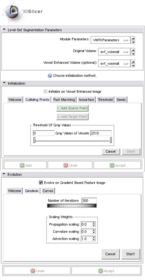
Usage
Installation
This module depends on the VmtkSlicerModule: see this page for installation notes.
The VMTKLevelSetSegmentation module can be installed as a 3D Slicer Extension or manually.
- Installation as a 3D Slicer Extension
Inside the 3D Slicer extension system, the module is called VMTKLevelSetSegmentation.
- Manual Installation
1. To get the latest source code, perform the following SVN checkout command:
svn checkout https://www.nitrc.org/svn/slicervmtklvlst
2. A directory named VMTKLevelSetSegmentation is downloaded.
3. Copy this directory to Slicer3-build/lib/Slicer3/Modules/ of your local Slicer installation.
When the module was successfully installed, it is available within 3D Slicer's module selector inside the category The Vascular Modeling Toolkit.
Examples, Use Cases & Tutorials
- Note use cases for which this module is especially appropriate, and/or link to examples.
- Link to examples of the module's use
- Link to any existing tutorials
Quick Tour of Features and Use
List all the panels in your interface, their features, what they mean, and how to use them. For instance:
- Input panel:
- Parameters panel:
- Output panel:
- Viewing panel:
Development
Dependencies
This module depends on the VMTK libraries which are provided in the VmtkSlicerModule. Therefore the VmtkSlicerModule has to be installed before the VMTKLevelSetSegmentation module can be used.
Known bugs
Follow this link to the VMTK in 3D Slicer bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
The complete source code is available at a NITRC SVN repository.
More Information
Acknowledgment
This work was funded by a grant of the Thomas-Gessmann Foundation part of the Founder Federation for German Science.
References
- Piccinelli M, Veneziani A, Steinman DA, Remuzzi A, Antiga L (2009) A framework for geometric analysis of vascular structures: applications to cerebral aneurysms. IEEE Trans Med Imaging. In press.
- Antiga L, Piccinelli M, Botti L, Ene-Iordache B, Remuzzi A and Steinman DA. An image-based modeling framework for patient-specific computational hemodynamics. Medical and Biological Engineering and Computing, 46: 1097-1112, Nov 2008.