Modules:UnbiasedNonLocalMeans-Documentation-3.6
Return to Slicer 3.6 Documentation
Module Name
UnbiasedNonLocalMeans
General Information
Module Type & Category
Type: Interactive
Category: CLI/DiffusionApplications
Authors, Collaborators & Contact
- Author: Antonio Tristán Vega , Santiago Aja Fernández
- Contact: atriveg@bwh.harvard.edu
Module Description
This module reduces noise (or unwanted detail) on a set of diffusion weighted images. For this, it filters the images using a Unbiased Non Local Means for Rician noise algorithm. It exploits not only the spatial redundancy, but the redundancy in similar gradient directions as well; it takes into account the N closest gradient directions to the direction being processed (a maximum of 5 gradient directions is allowed to keep a reasonable computational load, since we do not use neither similarity maps nor block-wise implementation).
The noise parameter is automatically estimated in the same way as in the jointLMMSE module.
A complete description of the algorithm may be found in:
Antonio Tristán-Vega and Santiago Aja-Fernández, "DWI filtering using joint information for DTI and HARDI", Medical Image Analysis, Volume 14, Issue 2, Pages 205-218. 2010.
Please, note that the execution of this filter is extremely slow, son only very conservative parameters (block size and search size as small as possible) should be used. Even so, its execution may take several hours. The advantage of this filter over joint LMMSE is its better preservation of edges and fine structures.
Usage
Examples, Use Cases & Tutorials
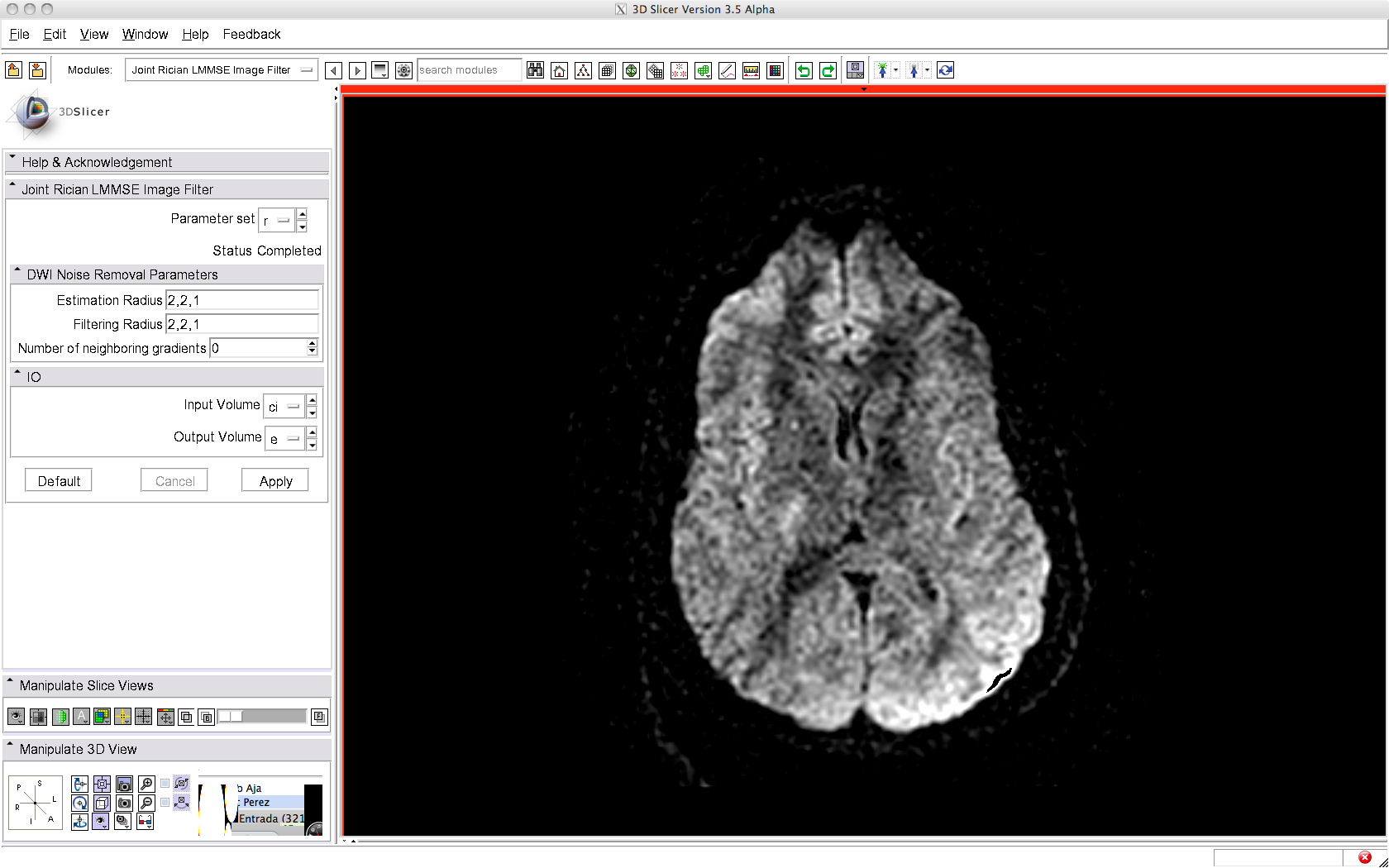
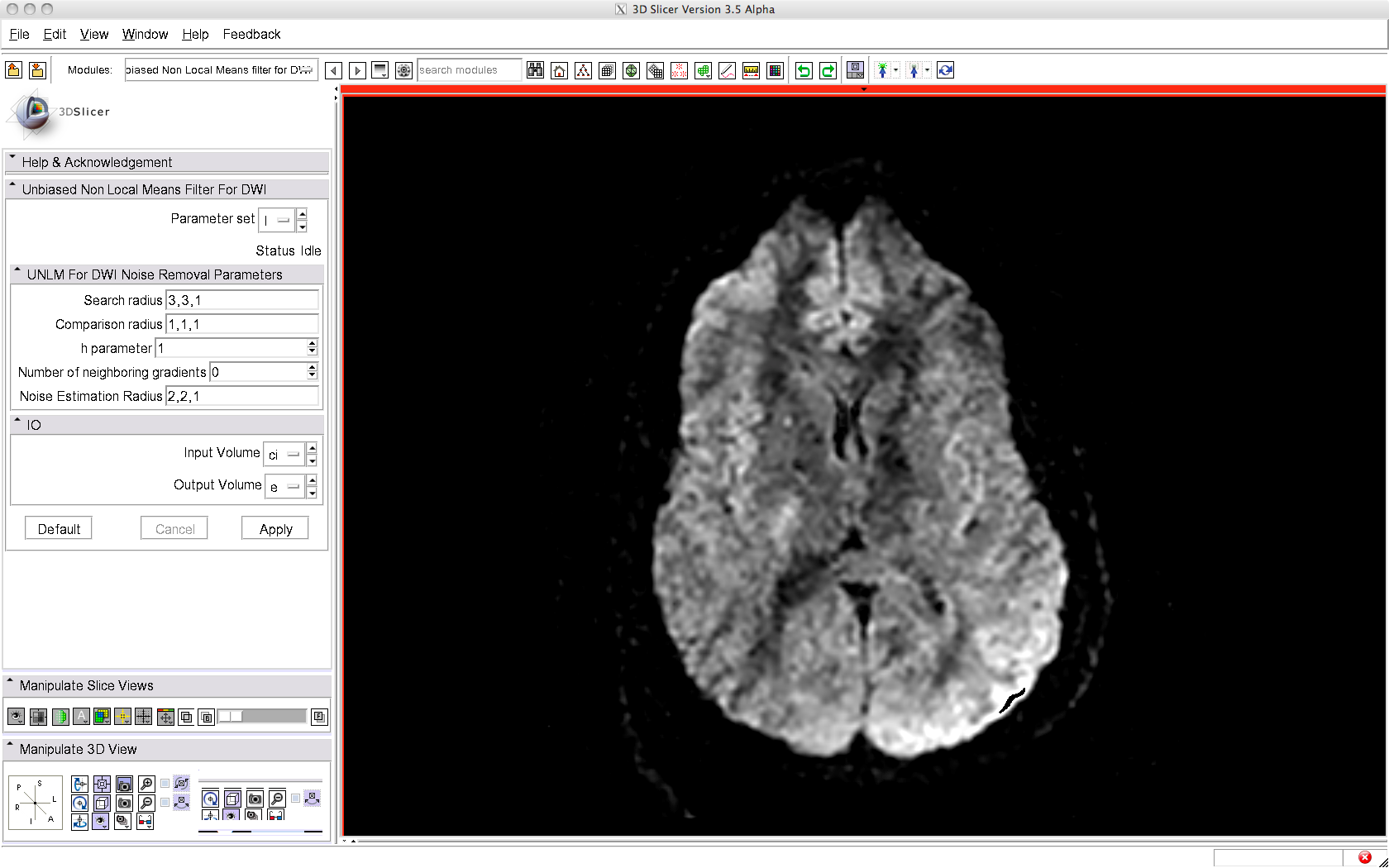
To use this filter, just load a DWI volume (gradient directions must be known) and apply the filter, as illustrated below. A brief summary of the parameters used is given below.
Before filtering:
After filtering:
Quick Tour of Features and Use
Just select a DWI, set the parameters (if you really need it), and you're ready to go.
- Input DWI Volume: set the DWI volume
- Output DWI Volume: the filtered DWI volume
- Estimation radius: This is the 3D radius of the neighborhood used for noise estimation. Noise power is estimated as the mode of the histogram of local variances
- Search radius: This filter works by computing the weighted average of pixels in a large neighborhood. The search radius is the 3D radius of that neighborhood.
- Comparison radius: The weights of the average are computed as the negative exponential of the (normalized) distance between two neighborhoods which have a 3D radius of this size.
- h: The normalization constant for the distance between neighborhoods. This parameter is related to the noise power, and should be in the range 0.8->1.2. Higher values produce more blurring (and more noise reduction), while lower values better preserve edges.
- Number of neighborhood gradients: This filter works gathering joint information from the N closest gradient directions to the one under study. This parameter is N. It is strongly recommended to keep this parameter less than 5 to keep a reasonable computational load. Even so, this filter is extremely slow.
Development
Dependencies
Volumes. Needed to load DWI volumes
Known bugs
Usability issues
This filter is very slow. May take hours.
Source code & documentation
Source Code: Follow this link
Doxygen documentation:
More Information
Acknowledgment
Antonio Tristan Vega, Santiago Aja Fernandez. University of Valladolid (SPAIN). Partially funded by grant number TEC2007-67073/TCM from the Ministerio de Ciencia e Innovación (Spain) and "FEDER" European Regional Development Fund.
References
- Antonio Tristán-Vega and Santiago Aja-Fernández, "DWI filtering using joint information for DTI and HARDI", Medical Image Analysis, Volume 14, Issue 2, Pages 205-218 (2010).
- Santiago Aja-Fernández, Antonio Tristán-Vega, and Carlos Alberola-López, "Noise estimation in single- and multiple-coil magnetic resonance data based on statistical models". Magnetic Resonance Imaging, Volume 27, Pages 1397-1409 (2009).