Difference between revisions of "Modules:SkullStripperModule"
| Line 49: | Line 49: | ||
===Usability issues=== | ===Usability issues=== | ||
| − | Developed and tested on T1 weighted images from OASIS database. Some limited test has been done on T2 weighted images. In some cases, when partial volume artifact is severe, the algorithm does not find the accurate brain boundary. | + | Developed and tested on T1 weighted images from OASIS database. Some limited test has been done on T2 weighted images. In some cases, when partial volume artifact is severe, the algorithm does not find the accurate brain boundary. This can be used for most of visualization tasks and applications where accuracy in cortex is not critical. |
===Source code & documentation=== | ===Source code & documentation=== | ||
Latest revision as of 15:04, 25 May 2010
Home < Modules:SkullStripperModuleReturn to Slicer 3.6 Documentation
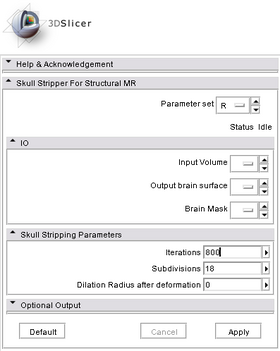
Module Name
Skull Stripper
|
|
General Information
Module Type & Category
Type: CLI
Category: Segmentation
Authors, Collaborators & Contact
- Author: Xiaodong Tao
- Contact: taox at research.ge.com
Module Description
Usage
Examples, Use Cases & Tutorials
Quick Tour of Features and Use
Development
Dependencies
No other modules are required for this module.
Known bugs
None.
Usability issues
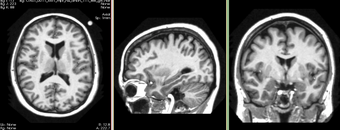
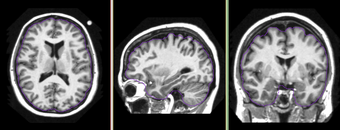
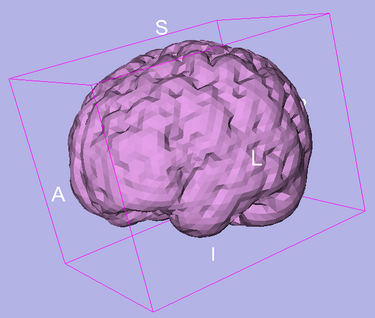
Developed and tested on T1 weighted images from OASIS database. Some limited test has been done on T2 weighted images. In some cases, when partial volume artifact is severe, the algorithm does not find the accurate brain boundary. This can be used for most of visualization tasks and applications where accuracy in cortex is not critical.
Source code & documentation
Source Code:
XML Description:
Usage:
More Information
Acknowledgment
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Implementation of the Fuzzy Classification was contributed by Dr. Ming-Ching Chang from GE Research.
OASIS datasets were used to generate images on this page.
References
- Xiaodong Tao, Ming-ching Chang, “A Skull Stripping Method Using Deformable Surface and Tissue Classification”, SPIE Medical Imaging, San Diego, CA, 2010.
- Ming-ching Chang, Xiaodong Tao “Subvoxel Segmentation and Representation of Brain Cortex Using Fuzzy Clustering and Gradient Vector Diffusion”, SPIE Medical Imaging, San Diego, CA, 2010.