Difference between revisions of "Modules:RobustStatisticsSeg-Documentation-3.6"
| Line 34: | Line 34: | ||
* This module is a general purpose segmentor. | * This module is a general purpose segmentor. | ||
* Link to examples of the module's use | * Link to examples of the module's use | ||
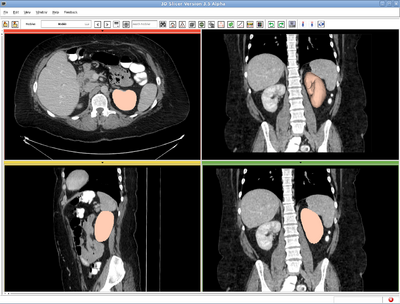
| − | Left kidney, CT image. IO time: 10sec, module running time: 12sec | + | Left kidney, CT image. IO time: 10sec, module running time: 12sec (Intel 3.0GHz) |
[[Image:RSSkidneyL.png | RSS left kidney result | 400px]] | [[Image:RSSkidneyL.png | RSS left kidney result | 400px]] | ||
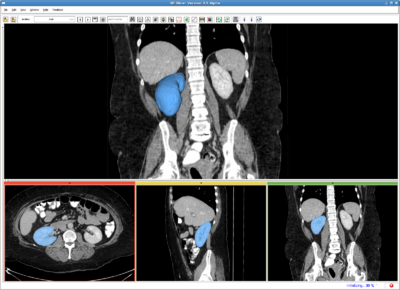
| − | Right kidney, CT image. IO time: 10sec, module running time: 15sec | + | Right kidney, CT image. IO time: 10sec, module running time: 15sec (Intel 3.0GHz) |
[[Image:RSS_rkidney.png | RSS right kidney result | 400px]] | [[Image:RSS_rkidney.png | RSS right kidney result | 400px]] | ||
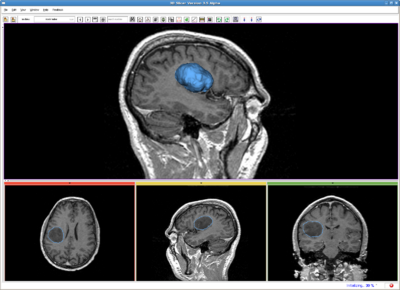
| − | Brain tumor, MR image. IO time: 3sec, Module running time: 2.5sec | + | Brain tumor, MR image. IO time: 3sec, Module running time: 2.5sec (Intel 3.0GHz) |
[[Image:RSS_tumor.png | RSS brain tumor result | 400px]] | [[Image:RSS_tumor.png | RSS brain tumor result | 400px]] | ||
Revision as of 16:29, 15 April 2010
Home < Modules:RobustStatisticsSeg-Documentation-3.6Return to Slicer 3.6 Documentation
Robust Statistics Segmentation
General Information
Module Type & Category
Type: CLI
Category: Segmentation
Authors, Collaborators & Contact
- Yi Gao (Author): Georgia Tech
- Allen Tannenbaum (Author): Georgia Tech
- Ron Kikinis (Author): BWH
- Contact: Yi Gao, yi.gao@gatech.edu
Module Description
This module is a general purpose segmenter. The target object is initialized by a label map. An active contour model then evolves to extract the desired boundary of the object.
Usage
Use Cases, Examples
- This module is a general purpose segmentor.
- Link to examples of the module's use
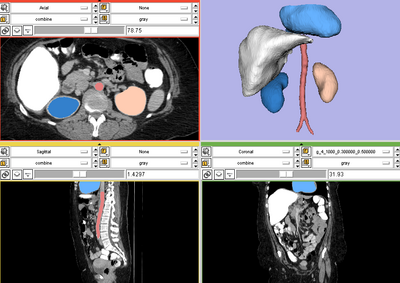
Left kidney, CT image. IO time: 10sec, module running time: 12sec (Intel 3.0GHz)
Right kidney, CT image. IO time: 10sec, module running time: 15sec (Intel 3.0GHz)
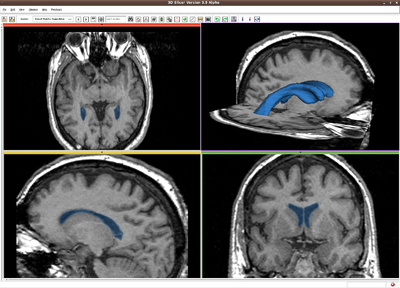
Brain tumor, MR image. IO time: 3sec, Module running time: 2.5sec (Intel 3.0GHz)
Tutorials
- First run:
- Give a rough estimate of the object volume and use the editing module to paint several non-zero labels, called seeds in the following, in the object.
- Run the module using the default parameters.
- Note:
- The Approximate volume is just a rough upper limit for the volume. It should be at least the size of the object. This is because when the volume reaches that, the program must stop. However, other criteria may stop the algorithm before the volume reaches this value.
- The positions of the seeds have to be in the object, preferably close to center.
- Troubleshooting
- Surface is too rough. Try:
- Increase "Boundary smoothness"
- Leakage into thin/narrow regions. Try:
- Increase "Boundary smoothness"
- leakage into similar (but still different) intensity regions (which is not necessarily thin), Try:
- Increase "Intensity homogeneity"
- Some regions are missed: Try (either one):
- Increase "Max volume"
- Decrease"Intensity homogeneity"
- Decrease "Boundary smoothness"
- Some regions are missed, at the same time leakages to some other regions. Try (either one)
- Increase "Intensity homogeneity"
- Add some other seeds
- Surface is too rough. Try:
- Data Set http://www.spl.harvard.edu/publications/bitstream/download/4217 (case3/grayscale.nrrd)
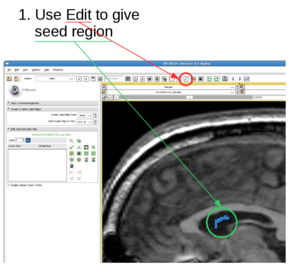
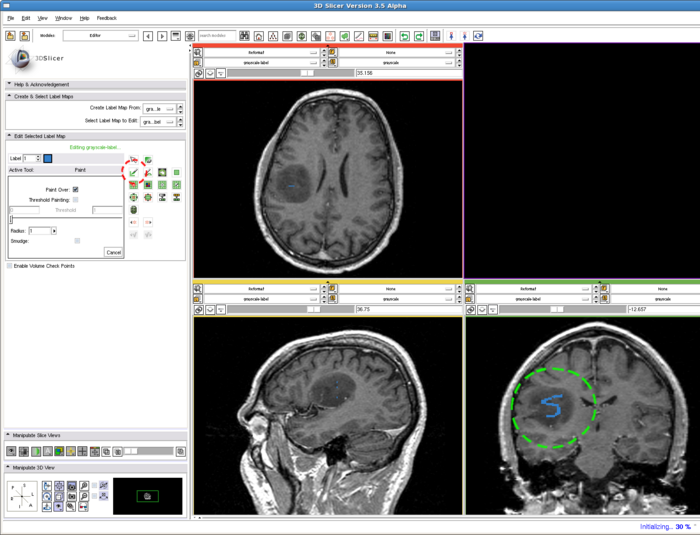
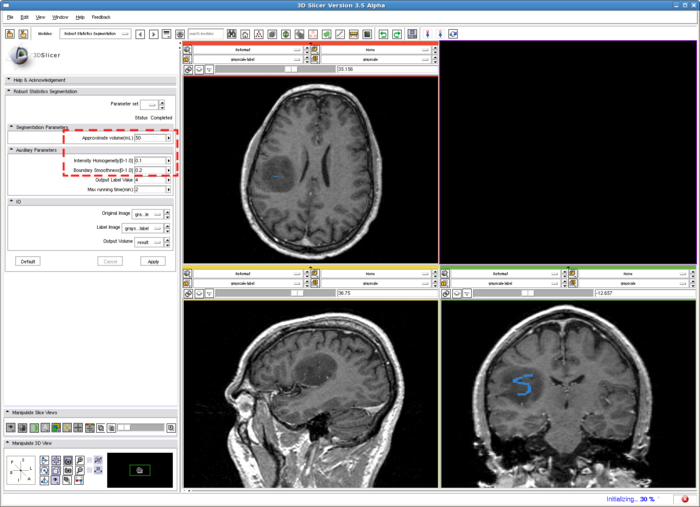
1. Draw label map in Editing module
In the editing module, select any drawing tool, for example the "Paint" tool circled in red. Draw some strokes in the target in one of the 2D views. In this example, we only freely draw the "S" shaped label circled in green.
2. Run RSS.
To run the RSS module. The parameters which may affect the segmentation results include: Approximate volume, Intensity homogeneity, Boundary smoothness, Max running time. The particular setting of the parameters for this example case is shown in the screenshot, in the red box. Some guidelines for adjusting them are given in the Troubleshooting section above, whereas their general roles in influencing the results are given in the Quick Tour of Features and Use section below. The sensitivity of the parameters are not quantitatively evaluated. In some easy cases, the algorithm is rather robust to the parameters. However for some objects with inhomogeneous intensity as well as irregular shape, the parameters may need to be carefully tuned.
- Multiple-value label map handling
The parameter "Output Label Value"(OLV) is for user to assign the output label value. Moreover, when user provided label map contains several labels, the target corresponding to the OLV is the one get segmented. More specifically, there are three difference situations:
- user provided label map contains only one label value, L. In this case, the output label value will be set to OLV, not matter what value L takes.
- user provided label map contains multiple label values, one of which matches OLV. Then only that label will be effective and all the others are discarded. The output will have label value OLV too.
- user provided label map contains multiple label values, but none matches OLV. Then all the non-zero labels will be considered as a single label value and then come back to the case 1 above.
In the current version, regardless of the number of different labels appearing in the label map, only one object is extracted from the image. The on-going work extends this to extracting multiple objects simultaneously.
Quick Tour of Features and Use
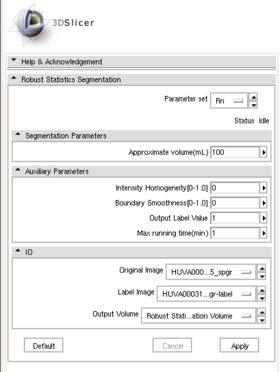
A list panels in the interface, their features, what they mean, and how to use them. For instance:
|
Development
Notes from the Developer(s)
Algorithms used, library classes depended upon, use cases, etc.
Dependencies
This module depends on the Slicer editing module, or outside input, to provide initial label image.
Tests
Test testing code and test data sets are included in this module to ensure its successful running on various platforms. The testing file is: SFLSRobustStat3DTestConsole.cxx, residing in the same directory as the module code.
Known bugs
No bug known on Apr 14.2010
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Links to the module's source code:
Source code:
Doxygen documentation:
More Information
Acknowledgment
This work was supported in part by grants from NSF, AFOSR, ARO, as well as by a grant from NIH (NAC P41 RR-13218) through Brigham and Women’s Hospital. An NSF Fellowship supported part of the work.