Difference between revisions of "Modules:ResampleVolume2-Documentation"
| Line 29: | Line 29: | ||
===Examples, Use Cases & Tutorials=== | ===Examples, Use Cases & Tutorials=== | ||
| − | * | + | * This module is especially appropriate when one has to realign an image to a reference image and already knows the transform that needs to be applied to its image. |
| − | |||
| − | |||
===Quick Tour of Features and Use=== | ===Quick Tour of Features and Use=== | ||
Revision as of 16:01, 15 September 2008
Home < Modules:ResampleVolume2-DocumentationReturn to Slicer Documentation
Module Name
MyModule
General Information
Module Type & Category
Type: CLI
Category: Filtering
Authors, Collaborators & Contact
- Francois Budin
- Sylvain Bouix, PNL
- Contact: Francois Budin , budin[at]bwh[dot]harvard[dot]edu
Module Description
This module implements image and vector-image resampling through the use of itk Transforms.It can also handle diffusion weighted MRI image resampling. 'Resampling' is performed in space coordinates, not pixel/grid coordinates. It is quite important to ensure that image spacing is properly set on the images involved. The interpolator is required since the mapping from one space to the other will often require evaluation of the intensity of the image at non-grid positions.
Usage
Examples, Use Cases & Tutorials
- This module is especially appropriate when one has to realign an image to a reference image and already knows the transform that needs to be applied to its image.
Quick Tour of Features and Use
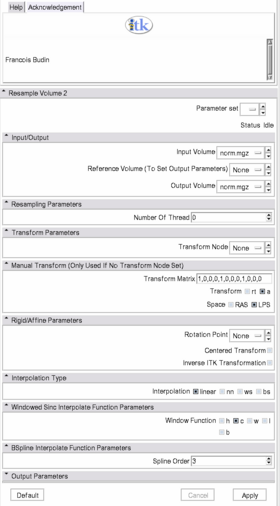
List all the panels in your interface, their features, what they mean, and how to use them. For instance:
- Input/Output: Defines input and output files.
- Input volume is the volume to resample,
- Reference Volume (optional) is the volume used to set the sampling parameters (spacing, orientation and dimensions),
- Output volume is the name of the output.
- Resampling Parameters: Sets the number of threads used to perform the resmapling.
- Transform Parameters: if the transform to be applied is in the slicer mrml tree, on can select it here.
- Manual Transform: if no tranform is set in previous panel, one can enter his own transform
- Transform Matrix: a 12 parameter affine transformation manually. The first 9 numbers represent a linear transformation matrix, the last 3 are a translation.
- Transform: forces the transform to be of rigid or affine type (affine is default)
- Space: specifies whether the matrix is expressed in LPS or RAS coordinate space.
- Rigid/Affine Parameters:
- Rotation Point: uses a fiducial to set a point around which the rotation defined in the transform needs to be performed.
- Centered Transform: sets the center of the transformation to the center of the image.
- Inverse ITK Transformation: inverses the transformation before applying it to the image
- Interpolation Type: sets the type of interpolation kernel to either linear, nearest neighbor (nn), windowed sinc (ws), or b-spline (bs).
- Windowed Sinc Interpolate Function Parameters: selects the type window function for the sinc interpolation h=hamming, c=cosine, w=welch, l=lanczos, b=blackman. This is only relevant if one selects ws as the interpolation type.
- BSpline Interpolate Function Parameters: The spline order.
- Output Parameters: if no reference volume is given, one can set the spacing, size, origin (as a fiducial), and direction matrix (also known as space directions) manually.
Development
Dependencies
Other modules or packages that are required for this module's use.
Known bugs
Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Customize following links for your module.
Links to documentation generated by doxygen.
More Information
Acknowledgment
Include funding and other support here.
References
Publications related to this module go here. Links to pdfs would be useful.